The Author ensures that the research has been conducted responsibly and ethically with adherence to all relevant regulations. read more..
- Plagarism Policy
- Peer Review Process
- Article Publishing Charges
- Grant Cover Letter
Join as Editor
Join as Associate Editor
- Join as Reviewer
- Manuscript Guidelines

Crimson Journals
Clinical research in animal science, --> modern research in dentistry --> <--.
- ISSN : 2770-6729
- Language of Publication : English
- Nature : Online
- [email protected] [email protected]
Journal Menu
- Classification
- Editorial Board
- Article In Press
- Current Issue
Latest News & Events
About Journal
Clinical Research in Animal Science is a peer-reviewed scholarly journal dedicated to the advancement in all fields of animal sciences. CRAS is a platform to advance the knowledge and to improve Animals health and are undergone with clinical trials to find new drug to alleviate pain. CRAS mainly focuses on providing an international forum to professionals, Researchers and other academicians to share the knowledge of scientific advancements through a peer reviewed publication on concerned subject area, it caters to the needs of health policy-makers, health managers, reflective practitioners and action-oriented researchers. CRAS main aim is to build a platform and communicate with researchers by publishing original research, reviews, case reports, opinion papers, research highlights, short communications and commentaries, etc. CRAS journal is dedicated to various disciplines of Animal sciences.
Latest Articles
Review Article

Progress of Research on Rabies and Rabies Vaccine
Published : July, 2024
Case Report
Homeopathic Treatment in a Puppy with Immune-Mediated Chronic Rheumatic Polyarthritis and Anemia due to Ehrlichia Sp. and Babesia Sp.
Published : March, 2024
Research Article
Identification of Genes related to Growth and Lipid Deposition from Transcriptome Profiles of Wujin Pigs and Landrace Pigs Muscle Tissue
Published : February, 2024
Short Communication
Evaluation Of Prophages and Anti- Microbial Resistance Profile of Bacillus Subtilis Using in Silico Approach
Published : January, 2024
The Rise of Integrative Veterinary Medicine: Use of Cam in Animal Healthcare
Published : December, 2023
Prevalence of Bovine Ticks in and Around Mendi Town, West Wollega, Oromia, Ethiopia
Histological and histochemical studies on the paraepiglottic tonsil of goats.
Published : November, 2023
Socio-Economic and Zootechnical Characterization of Fish Farming in the Noun Division (West Region- Cameroon)
Published : October, 2023
Mini Review
Is it Safe for Domesticated Animals to Drink Fresh Water in the Context of Fluoride Poisoning?
Published : September, 2023
Determination of Reproduction Performance Parameters of Sows Bred at the Pig Birth Centers of Giheta, Nyabunyegeri and Mahwa in Burundi
Published : August, 2023
Assessment on Community Knowledge, Attitude and Practice on Rabies in and Around Mendi Town, West Wollega, Oromia, Ethiopia
Some macro and micro elements in malapterurus electricus and gymnarchdae niloticus fish from khartoum fish market.
Published : July, 2023
Age Related Histological and Histochemical Studies on the Proventriculus of Broiler Chickens
Published : June, 2023
A Brief and Critical Review of Endemic Fluorosis in Domestic Animals of Scheduled Area of Rajasthan, India: Focus on Its Impact on Tribal Economy
Published : May, 2023
The Prognosis with Mechanical Ventilation in Dogs and Cats
Pubmed indexed articles.
- Dendrimer-Based Nanomedicine (Paramagnetic Nanoparticle, Nanocombretastatin, Nanocurcumin) for Glioblastoma Multiforme Imaging and Therapy PMID: 35237758
- Glioblastoma: Targeting Angiogenesis and Tyrosine Kinase Pathways PMID: 32924014
- The Conflict in East Ukraine: A Growing Need for Addiction Research and Substance Use Intervention for Vulnerable Populations PMID: 32363331
Track Your Article
Editor in chief.
Hirotada TSUJII
Ph.D in Agriculture from Faculty of Agriculture, Tohoku University
Approaches in Poultry, Dairy & Veterinary Sciences

Maria Kuman
Research Professor, PhD, Holistic Research Institute
Advances in Complementary & Alternative Medicine
Tomasz Karski
MD PhD, Professor, Vincent Pol University
Orthopedic Research Online Journal

Jiexiong Feng
Professor, Chief Doctor, Director of Department of Pediatric Surgery, Associate Director of Department of Surgery, Doctoral Supervisor Tongji hospital, Tongji medical college, Huazhong University of Science and Technology
Research in Pediatrics & Neonatology
Muhammad Atiqullah
Senior Research Engineer and Professor, Center for Refining and Petrochemicals, Research Institute, King Fahd University of Petroleum and Minerals (KFUPM), Dhahran, Saudi Arabia
Research & Development in Material Science

Ian James Martins
Fellow of International Agency for Standards and Ratings (IASR), Edith Cowan University, Sarich Neuroscience Research Institute
Advancements in Case Studies

Thomas F George
Chancellor Emeritus / Professor Emeritus of Chemistry and Physics, University of Missouri–St. Louis
Annals of Chemical Science Research
.jpg)
Jose Crisologo de Sales Silva
Ph.D in Science from the Federal University of Alagoas, UFAL, Brazil
Novel Research in Sciences

Naglaa Sami Adbel Aziz Mahmoud
Assistant Professor in College of Architecture, Art and Design
Academic Journal of Engineering Studies

Tong-Ching Tom Wu
Interim Dean, College of Education and Health Sciences, Director of Biomechanics Laboratory, Sport Science Innovation Program, Bridgewater State University
Research & Investigations in Sports Medicine

Dr. Jose Luis Turabian
Professor of numerous training courses in Family Medicine
Associative Journal of Health Sciences
Dariusz Jacek Jakóbczak
Assistant Professor, Department of Electronics and Computer Science
COJ Electronics & Communications

Önder Pekcan
Emeritus Professor of Physics, Kadir Has University, Turkey
Polymer Science: Peer Review Journal

Signup for Newsletter

Quick Links

Refer a Friend
Suggested By
Referrer Details

Advertise With Us
Our recent edition.

Modern Concepts & Developments in Agronomy

Environmental Analysis & Ecology Studies

Aspects in Mining & Mineral Science
Top Editors

Zhengcai Lou
Wenzhou Medical University, China

Fooyin University, Taiwan

Volkan Sarper Erikci
Saglik Bilimleri University, Turkey
Vincent Pol University, Poland
.jpg)
Thamil Selvam
National Defence University of Malaysia, Malaysia

Tarik Baykara
Dogus University, Turkey

Steven Smith
Hope College, USA

Stanislav Grigoriev
Russian Academy of Sciences, Russia

Southern Cross University, Australia

Shewikar Farrag
Umm Al-Qura University, Saudi Arabia

City University of New York, USA

Praveen K Maghelal
Khalifa University of Science & Technology, United Arab Emirates
Hebei Normal University, China
Nawal Mohamed Khalafallah
Alexandria University, Egypt

N K Kishore
Indian Institute of Technology Kharagpur, India

Muzzalupo Innocenzo
Council for Agriculture Research and Analysis of Agri Economy (CREA), Italy

King Fahd University of Petroleum and Minerals, Saudi Arabia

Mohamed A Rashed
King Abdulaziz University, Saudi Arabia

Maurice E Morgenstein
University of Oregon, USA

Martin Sweatman
University of Edinburgh, Scotland
.bmp)
University of Tennessee, USA
.jpg)
Manuel Velasco
Central University of Venezuela, Venezuela
.png)
Majid Monajjemi
Islamic Azad University Central Tehran Branch, Iran
.jpg)
Luisetto Mauro
Tourin University, Italy

Lloyd Arthur Jenkins
Teaching & Public Speaking, Spain
Leonardo Milella
Paeditric Hospital "Giovanni XXIII", Italy

Kanakis Dimitrios
University of Nicosia, Cyprus

Jose Luis Clua Espuny
Universidad Miguel Hernández de Elche, Spain

John Korstad
Oral Roberts University, USA

Jinliang Zhang
Beijing Normal University, China

Irina Koretsky
Howard University, USA
Edith Cowan University, Australia

Hamid Yahiya Hussain
Dubai Health Authority, UAE

Gundu HR Rao
University of Minnesota, USA
GP Karmakar

Ghassan George Haddad
Serhal Hospital, Lebanon

George Gregory Buttigieg
University of Malta, Malta

Fumihiko Hinoshita
National Center for Global Health and Medicine, Japan

Freida Pemberton
Molloy College, USA

Francisco Welington de Sousa Lima
Federal University of Piauí, Brazil

Florian Bert
Krankenhaus Nordwest Hospital, Germany
.png)
Fathi Habashi
Laval University, Canada

Dora Alicia Cortes Hernandez
Cinvestav-Unidad Saltillo, Mexico
.png)
Daniel Kinem
UPMC Hamot Neuroscience Institute, USA

Conxita Mestres Miralles
Ramon Llull University, Spain
Barry Kraynack
White Bear Associates, LLC, USA

Arkady S Voloshin
Lehigh University, USA

Alireza Heidari
California Southern University, USA
.png)
Alex Guskov
Institute of Solid State Physics of RAS, Russia

Alan Diego Briem Stamm
University of Buenos Aires, Argentina
Ahmed Nasr Ghanem
Mansoura University, Egypt

Afaf K El Ansary
King Saud University, Saudi Arabia

A Bernardes
University of Coimbra, Portugal
Financial Support

Latest e-Books
Latest video.

Identifiers
Linking ISSN (ISSN-L): 2770-6729
URL https://crimsonpublishers.com/cras/volume1-issue1-cras.php
Google https://www.google.com/search?q=ISSN+%222770-6729%22
Bing https://www.bing.com/search?q=ISSN+%222770-6729%22
Yahoo https://search.yahoo.com/search?p=ISSN%20%222770-6729%22
Library of Congress https://catalog.loc.gov/vwebv/search?searchCode=STNO&searchArg=2770-6729&searchType=1&limitTo=none&fromYear=&toYear=&limitTo=LOCA%3Dall&limitTo=PLAC%3Dall&limitTo=TYPE%3Dall&limitTo=LANG%3Dall&recCount=25
Resource information
Title proper: Clinical research in animal science.
Other variant title: Clinical researches in animal science
Other variant title: CRAS
Country: United States
Medium: Online
Record information
Last modification date: 17/11/2021
Type of record: Confirmed
ISSN Center responsible of the record: ISSN National Centre for the USA For all potential issues concerning the description of the publication identified by this bibliographic record (missing or wrong data etc.), please contact the ISSN National Centre mentioned above by clicking on the link.
downloads requested
Discover all the features of the complete ISSN records
Display mode x.
Labelled view
MARC21 view
UNIMARC view

CTAS is a peer-reviewed scholarly journal dedicated to the advancement in all fields of animal sciences. CRAS is a platform to advance the knowledge and to improve Animals health and are undergone with clinical trials to find new drug to alleviate pain.

- Search Menu
- Sign in through your institution
- Advance Articles
- High-Impact Collection
- Infographics
- Author Guidelines
- Submission Site
- Open Access
- Self-Archiving Policy
- Why Publish with Us?
- About Journal of Animal Science
- About the American Society of Animal Science
- Editorial Board
- Advertising & Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic

High-Impact Research from Journal of Animal Science
Explore a collection of the most read and most cited articles making an impact in Journal of Animal Science published within the past two years. This collection will be continuously updated with the journal's leading articles so be sure to revisit periodically to see what is being read and cited.
Also discover the articles being discussed the most on digital media by exploring this Altmetric report pulling the most discussed articles from the past year.

Recognizing and Celebrating Women in Science
Oxford University Press is proud to support diverse voices across our publishing. In this collection, we shine a spotlight on the representation of women in scientific fields, the gains that have been made in their fields, from research and major discoveries to advocacy and outreach, and amplify the voices of women who have made a career in scientific research.
Delve into the Women in Science collection
- Recommend to Your Librarian
- Advertising and Corporate Services
Affiliations
- Online ISSN 1525-3163
- Copyright © 2024 American Society of Animal Science
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
Loading metrics
Open Access
Essays articulate a specific perspective on a topic of broad interest to scientists.
See all article types »
A guide to open science practices for animal research
Contributed equally to this work with: Kai Diederich, Kathrin Schmitt
Affiliation German Federal Institute for Risk Assessment, German Centre for the Protection of Laboratory Animals (Bf3R), Berlin, Germany
* E-mail: [email protected]
- Kai Diederich,
- Kathrin Schmitt,
- Philipp Schwedhelm,
- Bettina Bert,
- Céline Heinl

Published: September 15, 2022
- https://doi.org/10.1371/journal.pbio.3001810
- Reader Comments
Translational biomedical research relies on animal experiments and provides the underlying proof of practice for clinical trials, which places an increased duty of care on translational researchers to derive the maximum possible output from every experiment performed. The implementation of open science practices has the potential to initiate a change in research culture that could improve the transparency and quality of translational research in general, as well as increasing the audience and scientific reach of published research. However, open science has become a buzzword in the scientific community that can often miss mark when it comes to practical implementation. In this Essay, we provide a guide to open science practices that can be applied throughout the research process, from study design, through data collection and analysis, to publication and dissemination, to help scientists improve the transparency and quality of their work. As open science practices continue to evolve, we also provide an online toolbox of resources that we will update continually.
Citation: Diederich K, Schmitt K, Schwedhelm P, Bert B, Heinl C (2022) A guide to open science practices for animal research. PLoS Biol 20(9): e3001810. https://doi.org/10.1371/journal.pbio.3001810
Copyright: © 2022 Diederich et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: The authors received no specific funding for this work.
Competing interests: I have read the journal’s policy and the authors of this manuscript have the following competing interests: All authors are employed at the German Federal Institute for Risk Assessment and part of the German Centre for the Protection of Laboratory Animals (Bf3R) which developed and hosts animalstudyregistry.org , a preregistration platform for animal studies and animaltestinfo.de, a database for non-technical project summaries (NTS) of approved animal study protocols within Germany.
Abbreviations: CC, Creative Commons; CIRS-LAS, critical incident reporting system in laboratory animal science; COVID-19, Coronavirus Disease 2019; DOAJ, Directory of Open Access Journals; DOI, digital object identifier; EDA, Experimental Design Assistant; ELN, electronic laboratory notebook; EU, European Union; IMSR, International Mouse Strain Resource; JISC, Joint Information Systems Committee; LIMS, laboratory information management system; MGI, Mouse Genome Informatics; NC3Rs, National Centre for the Replacement, Refinement and Reduction of Animals in Research; NTS, non-technical summary; RRID, Research Resource Identifier
Introduction
Over the past decade, the quality of published scientific literature has been repeatedly called into question by the failure of large replication studies or meta-analyses to demonstrate sufficient translation from experimental research into clinical successes [ 1 – 5 ]. At the same time, the open science movement has gained more and more advocates across various research areas. By sharing all of the information collected during the research process with colleagues and with the public, scientists can improve collaborations within their field and increase the reproducibility and trustworthiness of their work [ 6 ]. Thus, the International Reproducibility Networks have called for more open research [ 7 ].
However, open science practices have not been adopted to the same degree in all research areas. In psychology, which was strongly affected by the so-called reproducibility crisis, the open science movement initiated real practical changes leading to a broad implementation of practices such as preregistration or sharing of data and material [ 8 – 10 ]. By contrast, biomedical research is still lagging behind. Open science might be of high value for research in general, but in translational biomedical research, it is an ethical obligation. It is the responsibility of the scientist to transparently share all data collected to ensure that clinical research can adequately evaluate the risks and benefits of a potential treatment. When Russell and Burch published “The Principles of Humane Experimental Technique” in 1959, scientists started to implement their 3Rs principle to answer the ethical dilemma of animal welfare in the face of scientific progress [ 11 ]. By replacing animal experiments wherever possible, reducing the number of animals to a strict minimum, and refining the procedures where animals have still to be used, this ethical dilemma was addressed. However, in recent years, whether the 3Rs principle is sufficient to fully address ethical concerns about animal experiments has been questioned [ 12 ].
Most people tolerate the use of animals for scientific purposes only under the basic assumption that the knowledge gained will advance research in crucial areas. This implies that performed experiments are reported in a way that enables peers to benefit from the collected data. However, recent studies suggest that a large proportion of animal experiments are never actually published. For example, scientists working within the European Union (EU) have to write an animal study protocol for approval by the competent authorities of the respective country before performing an animal experiment [ 13 ]. In these protocols, scientists have to describe the planned study and justify every animal required for the project. By searching for publications resulting from approved animal study protocols from 2 German University Medical Centers, Wieschowski and colleagues found that only 53% of approved protocols led to a publication after 6 years [ 14 ]. Using a similar approach, Van der Naald and colleagues determined a publication rate of 60% at the Utrecht Medical Center [ 15 ]. In a follow-up survey, the respective researchers named so-called “negative” or null-hypothesis results as the main cause for not publishing outcomes [ 15 ]. The current scientific system is shaped by publishers, funders, and institutions and motivates scientists to publish novel, surprising, and positive results, revealing one of the many structural problems that the numerous efforts towards open science initiatives are targeting. Non-publication not only strongly contradicts ethical values, but also it compromises the quality of published literature by leading to overestimation of effect sizes [ 16 , 17 ]. Furthermore, publications of animal studies too often show poor reporting that strongly impairs the reproducibility, validity, and usefulness of the results [ 18 ]. Unfortunately, the idea that negative or equivocal findings can also contribute to the gain of scientific knowledge is frequently neglected.
So far, the scientific community using animals has shown limited resonance to the open science movement. Due to the strong controversy surrounding animal experiments, scientists have been reluctant to share information on the topic. Additionally, translational research is highly competitive and researchers tend to be secretive about their ideas until they are ready for publication or patent [ 19 , 20 ]. However, this missing openness could also point to a lack of knowledge and training on the many open science options that are available and suitable for animal research. Researchers have to be convinced of the benefits of open science practices, not only for science in general, but also for the individual researcher and each single animal. Yet, the key players in the research system are already starting to value open science practices. An increasing number of journals request open sharing of data, funders pay for open access publications and institutions consider open science practices in hiring decisions. Open science practices can improve the quality of work by enabling valuable scientific input from peers at the early stages of research projects. Furthermore, the extended communication that open science practices offer can draw attention to research and help to expand networks of collaborators and lead to new project opportunities or follow-up positions. Thus, open science practices can be a driver for careers in academia, particularly those of early career researchers.
Beyond these personal benefits, improving transparency in translational biomedical research can boost scientific progress in general. By bringing to light all the recorded research outputs that until now have remained hidden, the publication bias and the overestimation of effect sizes can be reduced [ 17 ]. Large-scale sharing of data can help to synthesize research outputs in preclinical research that will enable better decision-making for clinical research. Disclosing the whole research process will help to uncover systematic problems and support scientists in thoroughly planning their studies. In the long run, we predict that the implementation of open science practices will lead to the use of fewer animals in unintentionally repeated experiments that previously showed unreported negative results or in the establishment of methods by avoiding experimental dead ends that are often not published. More collaborations and sharing of materials and methods can further reduce the number of animal experiments used for the implementation of new techniques.
Open science can and should be implemented at each step of the research process ( Fig 1 ). A vast number of tools are already provided that were either directly conceptualized for animal research or can be adapted easily. In this Essay, we provide an overview of open science tools that improve transparency, reliability, and animal welfare in translational in vivo biomedical research by supporting scientists to clearly communicate their research and by supporting collaborative working. Table 1 lists the most prominent open science tools we discuss, together with their respective links. We have structured this Essay to guide you through which tools can be used at each stage of the research process, from planning and conducting experiments, through to analyzing data and communicating the results. However, many of these tools can be used at many different steps. Table 1 has been deposited on Zenodo and will be updated continuously [ 21 ].
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
Application of open science practices at each step of the research process can maximize the impact of performed animal experiments. The implementation of these practices will lead to less time pressure at the end of a project. Due to the connection of most of these open science practices, spending more time in the planning phase and during the conduction of experiments will save time during the data analysis and publication of the study. Indeed, consulting reporting guidelines early on, preregistering a statistical plan, and writing down crucial experimental details in an electronic lab notebook, will strongly accelerate the writing of a manuscript. If protocols or even electronic lab notebooks were made public, just citing these would simplify the writing of publications. Similarly, if a data management plan is well designed before starting data collection, analyzing, and depositing data in a public repository, as is increasingly required, will be fast. NTS, non-technical summary.
https://doi.org/10.1371/journal.pbio.3001810.g001
https://doi.org/10.1371/journal.pbio.3001810.t001
Planning the study
Transparent practices can be adopted at every stage of the research process. However, to ensure full effectivity, it is highly recommended to engage in detailed planning before the start of the experiment. This can prevent valuable time from being lost at the end of the study due to careless decisions being made at the beginning. Clarifying data management at the start of a project can help avoiding filing chaos that can be very time consuming to untangle. Keeping clear track of a project and study design will also help if new colleagues are included later on in the project or if entire project parts are handed over. In addition, all texts written on the rationale and hypothesis of the study or method descriptions, or design schemes created during the planning phase can be used in the final publications ( Fig 1 ). Similarly, information required for preregistration of animal studies or for reporting according to the ARRIVE guidelines are an extension of the details required for ethical approval [ 22 , 23 ]. Thus, the time burden within the planning phase is often overestimated. Furthermore, the thorough planning of experiments can avoid the unnecessary use of animals by preventing wrong avenues from being pursued.
Implementing open scientific practices at the beginning of a project does not mean that the idea and study plan must be shared immediately, but rather is critical for making the entire workflow transparent at the end of the project. However, optional early sharing of information can enable peers to give feedback on the study plan. Studies potentially benefit more from this a priori input than they would from the classical a posteriori peer-review process.
Most people perceive guidelines as advice that instructs on how to do something. However, it is sometimes useful to consider the term in its original meaning; “the line that guides us”. In this sense, following guidelines is not simply fulfilling a duty, but is a process that can help to design a sound research study and, as such, guidelines should be consulted at the planning stage of a project. The PREPARE guidelines are a list of important points that should be thought-out before starting a study involving animal experiments in order to reduce the waste of animals, promote alternatives, and increase the reproducibility of research and testing [ 24 ]. The PREPARE checklist helps to thoroughly plan a study and focuses on improving the communication and collaboration between all involved participants of the study (i.e., animal caretakers and scientists). Indeed, open science begins with the communication within a research facility. It is currently available in 33 languages and the responsible team from Norecopa, Norway’s 3R-center, takes requests for translations into further languages.
The UK Reproducibility Network has also published several guiding documents (primers) on important topics for open and reproducible science. These address issues such as data sharing [ 25 ], open access [ 26 ], open code and software [ 27 ], and preprints [ 28 ], as well as preregistration and registered reports [ 27 ]. Consultation of these primers is not only helpful in the relevant phases of the experiment but is also encouraged in the planning phase.
Although the ARRIVE guidelines are primarily a reporting guideline specifically designed for preparing a publication containing animal data, they can also support researchers when planning their experiments [ 22 , 23 ]. Going through the ARRIVE website, researchers will find tools and explanations that can support them in planning their experiments [ 29 ]. Consulting the ARRIVE checklist at the beginning of a project can help in deciding what details need to be documented during conduction of the experiments. This is particularly advisable, given that compliance to ARRIVE is still poor [ 18 ].
Experimental design
To maximize the validity of performed experiments and the knowledge gained, designing the study well is crucial. It is important that the chosen animal species reflects the investigated disease well and that basic characteristics of the animal, such as sex or age, are considered carefully [ 30 ]. The Canadian Institutes of Health Research provides a collection of resources on the integration of sex and gender in biomedical research with animals, including tips and tools for researchers and reviewers [ 31 ]. Additionally, it is advisable to avoid unnecessary standardization of biological and environmental factors that can reduce the external validity of results [ 32 ]. Meticulous statistical planning can further optimize the use of animals. Free to use online tools for calculating sample sizes such as G*Power or the inVivo software package for R can further support animal researchers in designing their statistical plan [ 33 , 34 ]. Randomization for the allocation of groups can be supported with specific tools for scientists like Research Randomizer, but also by simple online random number generators [ 35 ]. Furthermore, it might be advisable when designing the study to incorporate pathological analyses into the experimental plan. Optimal planning of tissue collection, performance of pathological procedures according to accepted best practices, and use of optimal pathological analysis and reporting methods can add some extra knowledge that would otherwise be lost. This can improve the reproducibility and quality of translational biomedicine, especially, but not exclusively, in animal studies with morphological endpoints. In all animal studies, unexpected deaths in experimental animals can occur and be the cause of lost data or missed opportunities to identify health problems [ 36 , 37 ].
To support researchers in designing their animal research, the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) has also developed the Experimental Design Assistant (EDA) [ 38 , 39 ]. This online tool helps researchers to better structure in vivo research by creating detailed schemes of the study design. It provides feedback on the entered design, drawing researcher’s attention to crucial decisions in the project. The resulting schemes can be used to transparently share the study design by uploading it into a study preregistration, enclosing it in a grant application, or submitting it with a final manuscript. The EDA can be used for different study designs in diverse scenarios and helps to communicate researcher plans to others [ 40 ]. The EDA might be particularly of interest to clarify very complex study designs involving multiple experimental groups. Working with the EDA might appear rather complex in the beginning, but the NC3R provides regular webinars that can help to answer any questions that arise.
Preregistration
Preregistration is an effective tool to improve the quality and transparency of research. To preregister their work, scientists must determine crucial details of the study before starting any experiment. Changes occurring during a study can be outlined at the end. A preregistered study plan should include at least the hypothesis and determine all the parameters that are known in advance. A description of the planned study design and statistical analysis will enable reviewers and peers to better retrace the workflow. It can prevent the intentional use of the flexibility of analysis to reach p -values under a certain significance level (e.g., p-hacking or HARKing (Hypothesizing After Results are Known)). With preregistration, scientists can also claim their idea at an early stage of their research with a citable individual identifier that labels the idea as their own. Some open preregistration platforms also provide a digital object identifier (DOI), which makes the registered study citable. Three public registries actively encourage the preregistration of animal studies conducted around the world: OSF registry, preclinicaltrials.eu, and animalstudyregistry.org [ 41 – 45 ]. Scientists can choose the registry according to their needs. Preregistering a study in a public registry supports scientists in planning their study and later to critically reevaluate their own work and assess its limitations and potentials.
As an alternative to public registries, researchers can also submit their study plan to one of hundreds of journals already publishing registered reports, including many journals open to animal research [ 8 ]. A submitted registered report passes 2 steps of peer review. In the first step, reviewers comment on the idea and the study design. After an “in-principle-acceptance,” researchers can conduct their study as planned. If the authors conduct the experiments as described in the accepted study protocol, the journal will publish the final study regardless of the outcome. This might be an attractive option, especially for early career researchers, as a manuscript is published at the beginning of a project with the guarantee of a future final publication.
The benefits of preregistration can already be observed in clinical research, where registration has been mandatory for most trials for more than 20 years. Preregistration in clinical research has helped to make known what has been tested and not just what worked and was published, and the implementation of trial registration has strongly reduced the number of publications reporting significant treatment effects [ 46 ]. In animal research, with its unrealistically high percentage of positive results, preregistration seems to be particularly worthwhile.
Research data management
To get the most out of performed animal experiments, effective sharing of data at the end of the study is essential. Sharing research data optimally is complex and needs to be prepared in advance. Thus, data management can be seen as one part of planning a study thoroughly. Many funders have recognized the value of the original research data and request a data management plan from applicants in advance [ 25 , 47 ]. Various freely available tools such as DMPTool or DMPonline already exist to design a research data management plan that complies to the requirements of different funders [ 48 , 49 ]. The data management plan defines the types of data collected and describes the handling and names responsible persons throughout the data lifecycle. This includes collecting the data, analyzing, archiving, and sharing it. Finally, a data management plan enables long-term access and the possibility for reuse by peers. Developing such a plan, whether it is required by funders or not, will later simplify the application of the FAIR data principle (see section on the FAIR data principle). The Longwood Medical Area Research Data Management Working Group from the Harvard Medical School developed a checklist to assist researchers in optimally managing their data throughout the data lifecycle [ 50 ]. Similarly, the Joint Information Systems Committee (JISC) provides a great research data management toolkit including a checklist for researchers planning their project [ 51 ]. Consulting this checklist in the planning phase of a project can prevent common errors in research data management.
Non-technical project summary
One instrument specifically conceived to create transparency on animal research for the general public is the so-called non-technical project summary (NTS). All animal protocols approved within the EU must be accompanied by these comprehensible summaries. NTSs are intended to inform the public about ongoing animal experiments. They are anonymous and include information on the objectives and potential benefits of the project, the expected harm, the number of animals, the species, and a statement of compliance with the requirements of the 3Rs principle. However, beyond simply informing the public, NTSs can also be used for meta-research to help identify new research areas with an increased need for new 3R technologies [ 52 , 53 ]. NTSs become an excellent tool to appropriately communicate the scientific value of the approved protocol and for meta-scientists to generate added value by systematically analyzing theses summaries if they fulfill a minimum quality threshold [ 54 , 55 ]. In 2021, the EU launched the ALURES platform ( Table 1 ), where NTSs from all member states are published together, opening the opportunities for EU-wide meta-research. NTSs are, in contrast to other open science practices, mandatory in the EU. However, instead of thinking of them as an annoying duty, it might be worth thoroughly drafting the NTS to support the goals of more transparency towards the public, enabling an open dialogue and reducing extreme opinions.
Conducting the experiments
Once the experiments begin, documentation of all necessary details is essential to ensure the transparency of the workflow. This includes methodological details that are crucial for replicating experiments, but also failed attempts that could help peers to avoid experiments that do not work in the future. All information should be stored in such a way that it can be found easily and shared later. In this area, many new tools have emerged in recent years ( Table 1 ). These tools will not only make research transparent for colleagues, but also help to keep track of one’s own research and improve internal collaboration.
Electronic laboratory notebooks
Electronic laboratory notebooks (ELNs) are an important pillar of research data management and open science. ELNs facilitate the structured and harmonized documentation of the data generation workflow, ensure data integrity, and keep track of all modifications made to the original data based on an audit trail option. Moreover, ELNs simplify the sharing of data and support collaborations within and outside the research group. Methodological details and research data become searchable and traceable. There is an extensive amount of literature providing advice on the selection and the implementation process of an ELN depending on the specific needs and research area and its discussion would be beyond the scope of this Essay [ 56 – 58 ]. Some ELNs are connected to a laboratory information management system (LIMS) that provides an animal module supporting the tracking of animal details [ 59 ]. But as research involving animals is highly heterogeneous, this might not be the only decision point and we cannot recommend a specific ELN that is suitable for all animal research.
ELNs are already established in the pharmaceutical industry and their use is on the rise among academics as well. However, due to concerns around costs for licenses, data security, and loss of flexibility, many research institutions still fear the expenses that the introduction of such a system would incur [ 56 ]. Nevertheless, an increasing number of academic institutions are implementing ELNs and appreciating the associated benefits [ 60 ]. If your institution already has an ELN, it might be easiest to just use the option available in the research environment. If not, the Harvard Medical School provides an extensive and updated overview of various features of different ELNs that can support scientists in choosing the appropriate one for their research [ 61 ]. There are many commercial ELN products, which may be preferred when the administrative workload should be outsourced to a large extent. However, open-source products such as eLabFTW or open BIS provide a greater opportunity for customization to meet specific needs of individual research institutions [ 62 – 64 ]. A huge number of options are available depending on the resources and the features required. Some scientists might prefer generic note taking tools such as Evernote or just a simple Word document that offers infinite flexibility, but specific ELNs can further support good record keeping practice by providing immutability, automated backups, standardized methods, and protocols to follow. Clearly defining the specific requirements expected might help to choose an adequate system that would improve the quality of the record compared to classical paper laboratory notebooks.
Sharing protocols
Adequate sharing of methods in translational biomedical sciences is key to reproducibility. Several repositories exist that simplify the publication and exchange of protocols. Writing down methods at the end of the project bears the risk that crucial details might be missing [ 65 ]. On protocols.io, scientists can note all methodological details of a procedure, complete them with uploaded documents, and keep them for personal use or share them with collaborators [ 66 ]. Authors can also decide at any point in time to make their protocol public. Protocols published on protocols.io receive a DOI and become citable; they can be commented on by peers and adapted according to the needs of the individual researcher. Protocol.io files from established protocols can also be submitted together with some context and sample datasets to PLOS ONE , where it can be peer-reviewed and potentially published [ 67 , 68 ]. Depending on the affiliation of the researchers to academia or industry and on an internal or public sharing of files, protocols.io can be free of charge or come with costs. Other journals also encourage their authors to deposit their protocols in a freely accessible repository, such as protocol exchange from Nature portfolio [ 69 ]. Another option might be to separately submit a protocol that was validated by its use in an already published research article to an online and peer-reviewed journal specific for research protocols, such as Bio-Protocol. A multitude of journals, including eLife and Science already collaborate with Bio-Protocol and recommend authors to publish the method in Bio-Protocol [ 70 ]. Bio-Protocol has no submission fees and is freely available to all readers. Both protocols.io and Bio-Protocol allow the illustration of complex scientific methods by uploading videos to published protocols. In addition, protocols can be deposited in a general research repository such as the Open Science Framework (OSF repository) and referenced in appropriate publications.

Sharing critical incidents
Sharing critical or even adverse events that occur in the context of animal experimentation can help other scientists to avoid committing the same mistakes. The system of sharing critical incidents is already established in clinical practice and helps to improve medical care [ 71 , 72 ]. The online platform critical incident reporting system in laboratory animal science (CIRS-LAS) represents the first preclinical equivalent to these clinical systems [ 73 ]. With this web-based tool, critical incidents in animal research can be reported anonymously without registration. An expert panel helps to analyze the incident to encourage an open dialogue. Critical incident reporting is still very marginal in animal research and performed procedures are very variable. These factors make a systemic analysis and a targeted search of incidence difficult. However, it may be of special interest for methods that are broadly used in animal research such as anesthesia. Indeed, a broad feed of this system with data on errors occurring in standard procedures today could help avoid critical incidences in the future and refine animal experiments.
Sharing animals, organs, and tissue
When we think about open science, sharing results and data are often in focus. However, sharing material is also part of a collaborative and open research culture that could help to greatly reduce the number of experimental animals used. When an animal is killed to obtain specific tissue or organs, the remainder is mostly discarded. This may constitute a wasteful practice, as surplus tissue can be used by other researchers for different analyses. More animals are currently killed as surplus than are used in experiments, demonstrating the potential for sharing these animals [ 74 , 75 ].
Sharing information on generated surplus is therefore not only economical, but also an effective way to reduce the number of animals used for scientific purposes. The open-source software Anishare is a straightforward way for breeders of genetically modified lines to promote their surplus offspring or organs within an institution [ 76 ]. The database AniMatch ( Table 1 ) connects scientists within Europe who are offering tissue or organs with scientists seeking this material. Scientists already sharing animal organs can support this process by describing it in publications and making peers aware of this possibility [ 77 ]. Specialized research communities also allow sharing of animal tissue or animal-derived products worldwide that are typically used in these fields on a collaborative basis via the SEARCH-framework [ 78 , 79 ]. Depositing transgenic mice lines into one of several repositories for mouse strains can help to further minimize efforts in producing new transgenic lines and most importantly reduce the number of surplus animals by supporting the cryoconservation of mouse lines. The International Mouse Strain Resource (IMSR) can be used to help find an adequate repository or to help scientists seeking a specific transgenic line find a match [ 80 ].
Analyzing the data
Animal researchers have to handle increasingly complex data. Imaging, electrophysiological recording, or automated behavioral tracking, for example, produce huge datasets. Data can be shared as raw numerical output but also as images, videos, sounds, or other forms from which raw numerical data can be generated. As the heterogeneity and the complexity of research data increases, infinite possibilities for analysis emerge. Transparently reporting how the data were processed will enable peers to better interpret reported results. To get the most out of performed animal experiments, it is crucial to allow other scientists to replicate the analysis and adapt it to their research questions. It is therefore highly recommended to use formats and tools during the analysis that allow a straightforward exchange of code and data later on.
Transparent coding
The use of non-transparent analysis codes have led to a lack of reproducibility of results [ 81 ]. Sharing code is essential for complex analysis and enables other researchers to reproduce results and perform follow-up studies, and citable code gives credit for the development of new algorithms ( Table 1 ). Jupyter Notebooks are a convenient way to share data science pipelines that may use a variety of coding languages, including like Python, R or Matlab, and also share the results of analyses in the form of tables, diagrams, images, and videos. Notebooks contain source code and can be published or collaboratively shared on platforms like GitHub or GitLab, where version control of source code is implemented. The data-archiving tool Zenodo can be used to archive a repository on GitHub and create a DOI for the archive. Thereby contents become citable. Using free and open-source programming language like R or Python will increase the number of potential researchers that can work with the published code. Best practice for research software is to publish the source code with a license that allows modification and redistribution.
Choice of data visualization
Choosing the right format for the visualization of data can increase its accessibility to a broad scientific audience and enable peers to better judge the validity of the results. Studies based on animal research often work with very small sample sizes. Visualizing these data in histograms may lead to an overestimation of the outcomes. Choosing the right dot plots that makes all recorded points visible and at the same time focusses on the summary instead of the individual points can further improve the intuitive understanding of a result. If the sample size is too low, it might not be meaningful to visualize error bars. A variety of freely available tools already exists that can support scientists in creating the most appropriate graphs for their data [ 82 ]. In particular, when representing microscopy results or heat maps, it should be kept in mind that a large part of the population cannot perceive the classical red and green representation [ 83 ]. Opting for the color-blind safe color maps and checking images with free tools such as color oracle ( Table 1 ) can increase the accessibility of graphs. Multiple journals have already addressed flaws in data visualization and have introduced new policies that will accelerate the uptake of transparent representation of results.
Publication of all study outcomes
Open science practices have received much attention in the past few years when it comes to publication of the results. However, it is important to emphasize that although open science tools have their greatest impact at the end of the project, good study preparation and sharing of the study plan and data early on can greatly increase the transparency at the end.
The FAIR data principle
To maximize the impact and outcome of a study, and to make the best long-term use of data generated through animal experiments, researchers should publish all data collected during their research according to the FAIR data principle. That means the data should be findable, accessible, interoperable, and reusable. The FAIR principle is thus an extension of open access publishing. Data should not only be published without paywalls or other access restrictions, but also in such a manner that they can be reused and further processed by others. For this, legal as well as technical requirements must be met by the data. To achieve this, the GoFAIR initiative has developed a set of principles that should be taken into account as early as at the data collection stage [ 49 , 84 ]. In addition to extensively described and machine-readable metadata, these principles include, for example, the application of globally persistent identifiers, the use of open file formats, and standardized communication protocols to ensure that humans and machines can easily download the data. A well-chosen repository to upload the data is then just the final step to publish FAIR data.
FAIR data can strongly increase the knowledge gained from performed animal experiments. Thus, the same data can be analyzed by different researchers and could be combined to obtain larger sample sizes, as already occurs in the neuroimaging community, which works with comparable datasets [ 85 ]. Furthermore, the sharing of data enables other researchers to analyze published datasets and estimate measurement reliabilities to optimize their own data collection [ 86 , 87 ]. This will help to improve the translation from animal research into clinics and simultaneously reduce the number of animal experiment in future.
Reporting guidelines
In preclinical research, the ARRIVE guidelines are the current state of the art when it comes to reporting data based on animal experiments [ 22 , 23 ]. The ARRIVE guidelines have been endorsed by more than 1,000 journals who ask that scientists comply with them when reporting their outcomes. Since the ARRIVE guidelines have not had the expected impact on the transparency of reporting in animal research publications, a more rigorous update has been developed to facilitate their application in practice (ARRIVE 2.0 [ 23 ]). We believe that the ARRIVE guidelines can be more effective if they are implemented at a very early stage of the project (see section on guidelines). Some more specialized reporting guidelines have also emerged for individual research fields that rely on animal studies, such as endodontology [ 88 ]. The equator network collects all guidelines and makes them easily findable with their search tool on their website ( Table 1 ). MERIDIAN also offers a 1-stop shop for all reporting guidelines involving the use of animals across all research sectors [ 89 ]. It is thus worth checking for new reporting guidelines before preparing a manuscript to maximize the transparency of described experiments.
Identifiers
Persistent identifiers for published work, authors, or resources are key for making public data findable by search engines and are thus a prerequisite for compliance to FAIR data principles. The most common identifier for publications will be a DOI, which makes the work citable. A DOI is a globally unique string assigned by the International DOI Foundation to identify content permanently and provide a persistent link to its location on the Internet. An ORCID ID is used as a personal persistent identifier and is recommendable to unmistakably identify an author ( Table 1 ). This will avoid confusions between authors with the same name or in the case of name changes or changes of affiliation. Research Resource Identifiers (RRID) are unique ID numbers that help to transparently report research resources. RRID also apply to animals to clearly identify the species used. RRID help avoid confusion between different names or changing names of genetic lines and, importantly, make them machine findable. The RRID Portal helps scientists find a specific RRID or create one if necessary ( Table 1 ). In the context of genetically altered animal lines, correct naming is key. The Mouse Genome Informatics (MGI) Database is the authoritative source of official names for mouse genes, alleles, and strains ([ 90 ]).
Preprint publication
Preprints have undergone unprecedented success, particularly during the height of the Coronavirus Disease 2019 (COVID-19) pandemic when the need for rapid dissemination of scientific knowledge was critical. The publication process for scientific manuscripts in peer-reviewed journals usually requires a considerable amount of time, ranging from a few months to several years, mainly due to the lengthy review process and inefficient editorial procedures [ 91 , 92 ]. Preprints typically precede formal publication in scientific journals and, thus, do not go through a peer review process, thus, facilitating the prompt open dissemination of important scientific findings within the scientific community. However, submitted papers are usually screened and checked for plagiarism. Preprints are assigned a DOI so they can be cited. Once a preprint is published in a journal, its status is automatically updated on the preprint server. The preprint is linked to the publication via CrossRef and mentioned accordingly on the website of the respective preprint platform.
After initial skepticism, most publishers now allow papers to be posted on preprint servers prior to submission. An increasing number of journals even allow direct submission of a preprint to their peer review process. The US National Institutes of Health and the Wellcome Trust, among other funders, also encourage prepublication and permit researchers to cite preprints in their grant applications. There are now numerous preprint repositories for different scientific disciplines. BioASAP provides a searchable database for preprint servers that can help in identifying the one that best matches an individual’s needs [ 93 ]. The most popular repository for animal research is bioRxiv, which is hosted by the Cold Spring Harbor Laboratory ( Table 1 ).
The early exchange of scientific results is particularly important for animal research. This acceleration of the publication process can help other scientists to adapt their research or could even prevent animal experiments if other scientists become aware that an experiment has already been done before starting their own. In addition, preprints can help to increase the visibility of research. Journal articles that have a corresponding preprint publication have higher citation and Altmetric counts than articles without preprint [ 94 ]. In addition, the publication of preprints can help to combat publication bias, which represents a major problem in animal research [ 16 ]. Since journals and readers prioritize cutting-edge studies with positive results over inconclusive or negative results, researchers are reluctant to invest time and money in a manuscript that is unlikely to be accepted in a high-impact journal.
In addition to the option of publishing as preprint, other alternative publication formats have recently been introduced to facilitate the publication of research results that are hard to publish in traditional peer-reviewed journals. These include micro publications, data repositories, data journals, publication platforms, and journals that focus on negative or inconclusive results. The tool fiddle can support scientists in choosing the right publication format [ 95 , 96 ].
Open access publication
Publishing open access is one of the most established open science strategies. In contrast to the FAIR data principle, the term open access publication refers usually to the publication of a manuscript on a platform that is accessible free of charge—in translational biomedical research, this is mostly in the form of a scientific journal article. Originally, publications accessible free of charge were the answer to the paywalls established by renowned publishing houses, which led to social inequalities within and outside the research system. In translational biomedical research, the ethical aspect of urgently needed transparency is another argument in favor of open access publication, as these studies will not only be findable, but also internationally readable.
There are different ways of open access publishing; the 2 main routes are gold open access and green open access. Numerous journals offer now gold open access. It refers to the immediate and fully accessible publication of an article. The Directory of Open Access Journals (DOAJ) provides a complete and updated list for high-quality, open access, and peer-reviewed journals [ 97 ]. Charité–Universitätsmedizin Berlin offers a specific tool for biomedical open access journals that supports animal researchers to choose an appropriate journal [ 49 ]. In addition, the Sherpa Romeo platform is a straightforward way to identify publisher open access policies on a journal-by-journal basis, including information on preprints, but also on licensing of articles [ 51 ]. Hybrid open access refers to openly accessible articles in otherwise paywalled journals. By contrast, green open access refers to the publication of a manuscript or article in a repository that is mostly operated by institutions and/or universities. The publication can be exclusively on the repository or in combination with a publisher. In the quality-assured, global Directory of Open Access Repositories (openDOAR), scientists can find thousands of indexed open access repositories [ 49 ]. The publisher often sets an embargo during which the authors cannot make the publication available in the repository, which can restrict the combined model. It is worth mentioning that gold open access is usually more expensive for the authors, as they have to pay an article processing charge. However, the article’s outreach is usually much higher than the outreach of an article in a repository or available exclusively as subscription content [ 98 ]. Diamond open access refers to publications and publication platforms that can be read free of charge by anyone interested and for which no costs are incurred by the authors either. It is the simplest and fairest form of open access for all parties involved, as no one is prevented from participating in scientific discourse by payment barriers. For now, it is not as widespread as the other forms because publishers have to find alternative sources of revenue to cover their costs.
As social media and the researcher’s individual public outreach are becoming increasingly important, it should be remembered that the accessibility of a publication should not be confused with the licensing under which the publication is made available. In order to be able to share and reuse one’s own work in the future, we recommend looking for journals that allow publications under the Creative Commons licenses CC BY or CC BY-NC. This also allows the immediate combination of gold and green open access.
Creative commons licenses
Attributing Creative Commons (CC) licenses to scientific content can make research broadly available and clearly specifies the terms and conditions under which people can reuse and redistribute the intellectual property, namely publications and data, while giving the credit to whom it deserves [ 49 ]. As the laws on copyright vary from country to country and law texts are difficult to understand for outsiders, the CC licenses are designed to be easily understandable and are available in 41 languages. This way, users can easily avoid accidental misuse. The CC initiative developed a tool that enables researchers to find the license that best fits their interests [ 49 ]. Since the licenses are based on a modular concept ranging from relatively unrestricted licenses (CC BY, free to use, credit must be given) to more restricted licenses (CC BY-NC-ND, only free to share for non-commercial purposes, credit must be given), one can find an appropriate license even for the most sensitive content. Publishing under an open CC license will not only make the publication easy to access but can also help to increase its reach. It can stimulate other researchers and the interested public to share this article within their network and to make the best future use of it. Bear in mind that datasets published independently from an article may receive a different CC license. In terms of intellectual property, data are not protected in the same way as articles, which is why the CC initiative in the United Kingdom recommends publishing them under a CC0 (“no rights reserved”) license or the Public Domain Mark. This gives everybody the right to use the data freely. In an animal ethics sense, this is especially important in order to get the most out of data derived from animal experiments.
Data and code repositories
Sharing research data is essential to ensure reproducibility and to facilitate scientific progress. This is particularly true in animal research and the scientific community increasingly recognizes the value of sharing research data. However, even though there is increasing support for the sharing of data, researchers still perceive barriers when it comes to doing so in practice [ 99 – 101 ]. Many universities and research institutions have established research data repositories that provide continuous access to datasets in a trusted environment. Many of these data repositories are tied to specific research areas, geographic regions, or scientific institutions. Due to the growing number and overall heterogeneity of these repositories, it can be difficult for researchers, funding agencies, publishers, and academic institutions to identify appropriate repositories for storing and searching research data.
Recently, several web-based tools have been developed to help in the selection of a suitable repository. One example is Re3data, a global registry of research data repositories that includes repositories from various scientific disciplines. The extensive database can be searched by country, content (e.g., raw data, source code), and scientific discipline [ 49 ]. A similar tool to help find a data archive specific to the field is FAIRsharing, based at Oxford University [ 102 ]. If there is no appropriate subject-specific data repository or one seems unsuitable for the data, there are general data repositories, such as Open Science Framework, figshare, Dryad, or Zenodo. To ensure that data stored in a repository can be found, a DOI is assigned to the data. Choosing the right license for the deposited code and data ensures that authors get credit for their work.
Publication and connection of all outcomes
If scientists have used all available open science tools during the research process, then publishing and linking all outcomes represents the well-deserved harvest ( Fig 2 ). At the end of a research process, researchers will not just have 1 publication in a journal. Instead, they might have a preregistration, a preprint, a publication in a journal, a dataset, and a protocol. Connecting these outcomes in a way that enables other scientists to better assess the results that link these publications will be key. There are many examples of good open science practices in laboratory animal science, but we want to highlight one of them to show how this could be achieved. Blenkuš and colleagues investigated how mild stress-induced hyperthermia can be assessed non-invasively by thermography in mice [ 103 ]. The study was preregistered with animalstudyregistry.org , which is referred to in their publication [ 104 ]. A deviation from the originally preregistered hypothesis was explained in the manuscript and the supplementary material was uploaded to figshare [ 105 ].
Application of open science practices can increase the reproducibility and visibility of a research project at the same time. By publishing different research outputs with more detailed information than can be included in a journal article, researchers enable peers to replicate their work. Reporting according to guidelines and using transparent visualization will further improve this reproducibility. The more research products that are generated, the more credit can be attributed. By communicating on social media or additionally publishing slides from delivered talks or posters, more attention can be raised. Additionally, publishing open access and making the work machine-findable makes it accessible to an even broader number of peers.
https://doi.org/10.1371/journal.pbio.3001810.g002
It might also be helpful to provide all resources from a project in a single repository such as Open Science Framework, which also implements other, different tools that might have been used, like GitHub or protocols.io.
Communicating your research
Once all outcomes of the project are shared, it is time to address the targeted peers. Social media is an important instrument to connect research communities [ 106 ]. In particular, Twitter is an effective way to communicate research findings or related events to peers [ 107 ]. In addition, specialized platforms like ResearchGate can support the exchange of practical experiences ( Table 1 ). When all resources related to a project are kept in one place, sharing this link is a straightforward way to reach out to fellow scientists.
With the increasing number of publications, science communication has become more important in recent years. Transparent science that communicates openly with the public contributes to strengthening society’s trust in research.
Conclusions
Plenty of open science tools are already available and the number of tools is constantly growing. Translational biomedical researchers should seize this opportunity, as it could contribute to a significant improvement in the transparency of research and fulfil their ethical responsibility to maximize the impact of knowledge gained from animal experiments. Over and above this, open science practices also bear important direct benefits for the scientists themselves. Indeed, the implementation of these tools can increase the visibility of research and becomes increasingly important when applying for grants or in recruitment decisions. Already, more and more journals and funders require activities such as data sharing. Several institutions have established open science practices as evaluation criteria alongside publication lists, impact factor, and h-index for panels deciding on hiring or tenure [ 108 ]. For new adopters, it is not necessary to apply all available practices at once. Implementing single tools can be a safe approach to slowly improve the outreach and reproducibility of one’s own research. The more open science products that are generated, the more reproducible the work becomes, but also the more the visibility of a study increases ( Fig 2 ).
As other research fields, such as social sciences, are already a step ahead in the implementation of open science practices, translational biomedicine can profit from their experiences [ 109 ]. We should thus keep in mind that open science comes with some risks that should be minimized early on. Indeed, the more open science practices become incentivized, the more researchers could be tempted to get a transparency quality label that might not be justified. When a study is based on a bad hypothesis or poor statistical planning, this cannot be fixed by preregistration, as prediction alone is not sufficient to validate an interpretation [ 110 ]. Furthermore, a boom of data sharing could disconnect data collectors and analysts, bearing the risk that researchers performing the analysis lack understanding of the data. The publication of datasets could also promote a “parasitic” use of a researcher’s data and lead to scooping of outcomes [ 111 ]. Stakeholders could counteract such a risk by promoting collaboration instead of competition.
During the COVID-19 pandemic, we have seen an explosion of preprint publications. This unseen acceleration of science might be the adequate response to a pandemic; however, the speeding up science in combination with the “publish or perish” culture could come at the expense of the quality of the publication. Nevertheless, a meta-analysis comparing the quality of reporting between preprints and peer-reviewed articles showed that the quality of reporting in preprints in the life sciences is at most slightly lower on average compared to peer-reviewed articles [ 112 ]. Additionally, preprints and social media have shown during this pandemic that a premature and overconfident communication of research results can be overinterpreted by journalists and raise unfounded hopes or fears in patients and relatives [ 113 ]. By being honest and open about the scope and limitations of the study and choosing communication channels carefully, researchers can avoid misinterpretation. It should be noted, however, that by releasing all methodological details and data in research fields such as viral engineering, where a dual use cannot be excluded, open science could increase biosecurity risk. Implementing access-controlled repositories, application programming interfaces, and a biosecurity risk assessment in the planning phase (i.e., by preregistration) could mitigate this threat [ 114 ].
Publishing in open access journals often involves higher publication costs, which makes it more difficult for institutes and universities from low-income countries to publish there [ 115 ]. Equity has been identified as a key aim of open science [ 116 ]. It is vital, therefore, that existing structural inequities in the scientific system are not unintentionally reinforced by open science practices. Early career researchers have been the main drivers of the open science movement in other fields even though they are often in vulnerable positions due to short contracts and hierarchical and strongly networked research environments. Supporting these early career researchers in adopting open science tools could significantly advance this change in research culture [ 117 ]. However, early career researchers can already benefit by publishing registered reports or preprints that can provide a publication much faster than conventional journal publications. Communication in social media can help them establish a network enabling new collaborations or follow-up positions.
Even though open science comes with some risks, the benefits easily overweigh these caveats. If a change towards more transparency is accompanied by the implementation of open science in the teaching curricula of the universities, most of the risks can be minimized [ 118 ]. Interestingly, we have observed that open science tools and infrastructure that are specific to animal research seem to mostly come from Europe. This may be because of strict regulations within Europe for animal experiments or because of a strong research focus in laboratory animal science along with targeted research funding in this region. Whatever the reason might be, it demonstrates the important role of research policy in accelerating the development towards 3Rs and open science.
Overall, it seems inevitable that open science will eventually prevail in translational biomedical research. Scientists should not wait for the slow-moving incentive framework to change their research habits, but should take pioneering roles in adopting open science tools and working towards more collaboration, transparency, and reproducibility.
Acknowledgments
The authors gratefully acknowledge the valuable input and comments from Sebastian Dunst, Daniel Butzke, and Nils Körber that have improved the content of this work.
- View Article
- PubMed/NCBI
- Google Scholar
- 6. Cary Funk MH, Brian Kennedy, Courtney Johnson. Americans say open access to data and independent review inspire more trust in research findings. Pew Research Center Website: Pew Research Center; 2019. Available from: https://www.pewresearch.org/science/2019/08/02/americans-say-open-access-to-data-and-independent-review-inspire-more-trust-in-research-findings/ .
- 7. International Reproducibility Networks. International Networks Statement UK Reproducibility Network Website: UK Reproducibility Network. 2021. Available from: https://cpb-eu-w2.wpmucdn.com/blogs.bristol.ac.uk/dist/b/631/files/2021/09/International-Networks-Statement-v1.0.pdf .
- 13. Article 36 of Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 amended by Regilation (EU) 2019/1010 of the European Parliament and of the Council of 5 June 2019. OJEU. 2010;L276:36.
- 19. American Association for Cancer Research. Editorial Policies. 2021. Available from: https://aacrjournals.org/content/authors/editorial-policies .
- 21. Diederich K, Schmitt K, Schwedhelm P, Bert B, Heinl C. Open Science Toolbox for Animal Research. Zenodo. 2022. Available from: https://zenodo.org/record/6497560 .
- 29. NC3R. ARRIVE guidelines. NC3R Website. Available from: https://arriveguidelines.org/ .
- 31. Canadian Institutes of Health Research. How to integrate sex and gender into research. Website of the Canadian Institutes of Health Research: Canadian Institutes of Health Research. 2019 [cited 2019 Aug 21]. Available from: https://cihr-irsc.gc.ca/e/50836.html .
- 33. Simon T, Bate RAC. InVivoStat. Available from: https://invivostat.co.uk/ .
- 35. Urbaniak G, Plous S. Research randomizer (version 4.0) [computer software]. 2013.
- 47. Medical Research Council’s. Data sharing policy. UK Research and Innovation Website 2021. Available from: https://www.ukri.org/publications/mrc-data-sharing-policy/ .
- 48. University of California Curation Center. DMPTool. 2021. Available from: https://dmptool.org/ .
- 49. Digital Curation Centre. DMPOnline. Available from: https://dmponline.dcc.ac.uk/ . Digital Curation Centre; 2021.
- 50. Harvard Longwood Medical Area Research Data Management Working Group. Biomedical Data Lifecycle. Harvard Medical School Website: Harvard Medical School; 2021. Available from: https://datamanagement.hms.harvard.edu/about/what-research-data-management/biomedical-data-lifecycle .
- 51. Joint Information Systems Committee. Research data management toolkit JISC Website: JISC; 2018. Available from: https://www.jisc.ac.uk/guides/rdm-toolkit .
- 54. German Centre for the Protection of Laboratory Animals (Bf3R). NTPs—Nicht Technische Projektzusammenfassungen 3R-SMART; 2020. Available from: https://www.3r-smart.de/index.php?id=6895 .
- 55. Understanding Animal Research. Guide to writing non-technical summaries concordat on openness on animal research in the UK2018. Available from: https://concordatopenness.org.uk/guide-to-writing-non-technical-summaries .
- 56. Gerlach B, Untucht C, Stefan A. Electronic Lab Notebooks and Experimental Design Assistants. In: Bespalov A, Michel MC, Steckler T, editors. Good Research Practice in Non-Clinical Pharmacology and Biomedicine. Cham: Springer International Publishing; 2020. p. 257–75.
- 58. Adam BL, Birte L. ELN Guide: electronic laboratory notebooks in the context of research data management and good research practice–a guide for the life sciences. Cologne, Germany: ZB MED–Information Centre for Life Sciences; 2021.
- 59. AgileBio. LabCollector Website https://labcollector.com/labcollector-lims/features/modules/animals-module/2022 . Available from: https://labcollector.com/labcollector-lims/features/modules/animals-module/ .
- 61. Harvard Longwood Medical Area Research Data Management Working Group. Electronic Lab Notebook Comparison Matrix. Zenodo. 2021.
- 70. Bio-protocol. Collaborating Journals bio-protocol website2021. Available from: https://bio-protocol.org/default.aspx?dw=Collaborating .
- 76. Dinkel H. anishare: GitHub; [updated June 2018]. Available from: https://github.com/hdinkel/anishare .
- 89. O’Connor AM. MERIDIAN: Menagerie of Reporting guidelines Involving Animals. Iowa State University; 2022. Available from: https://meridian.cvm.iastate.edu/ .
- 90. The Jackson Laboratory. Mouse Nomenclature Home Page at the Mouse Genome Informatics website World Wide Web: The Jackson Laboratory,Bar Harbor, Maine. Available from: http://www.informatics.jax.org/mgihome/nomen/index.shtml .
- 97. Directory of Open Access Journals. Find open access journals & articles. Available from: https://doaj.org/ . Directory of Open Access Journals, [DOAJ]; 2021.
- 98. Gold Open Access research has greater societal impact as used more outside of academia [press release]. Springer Nature Website: Springer. Nature. 2020;30:2020.
- 104. Franco NH. Can we use infrared thermography for assessing emotional states in mice?—A comparison between handling-induced stress by different techniques. Available from: animalstudyregistry.org . German Federal Institute for Risk Assessment (BfR); 2020. https://doi.org/10.17590/asr.0000224
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Author Biographies
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
The role of behavioral management in enhancing clinical care and efficiency, minimizing social disruption, and promoting welfare in captive primates.

Simple Summary
1. introduction, 2. materials and methods, 2.1. animal subjects, 2.2. behavioral management program, 2.3. iv access for blood sampling and animal monitoring, 2.4. statistical analysis, 3.1. behavioral management impact on intervention duration, 3.2. behavioral management impact on medical intervention and recovery (total duration), 3.3. side effects, 3.4. programmatic impact and total animal burden, 3.5. application of behavioral management for veterinary care—case studies, 3.5.1. case 1—wound management, 3.5.2. case 2—arthritis, 3.5.3. case 3—acute and supportive care, 4. discussion, 5. conclusions, author contributions, institutional review board statement, informed consent statement, data availability statement, acknowledgments, conflicts of interest.
- Parker, J.C.; Smarr, K.L.; Buckelew, S.P.; Stucky-ropp, R.C.; Hewett, J.E.; Johnson, J.C.; Wright, G.E.; Irvin, W.S.; Walker, S.E. Effects of stress management on clinical outcomes in rheumatoid arthritis. Arthritis Rheum. 1995 , 38 , 1807–1818. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Gouin, J.-P.; Kiecolt-Glaser, J.K. The impact of psychological stress on wound healing: Methods and mechanisms. Immunol. Allergy Clin. 2011 , 31 , 81–93. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- DiMatteo, M.R.; Lepper, H.S.; Croghan, T.W. Depression is a risk factor for noncompliance with medical treatment: Meta-analysis of the effects of anxiety and depression on patient adherence. Arch. Intern. Med. 2000 , 160 , 2101–2107. [ Google Scholar ] [ CrossRef ]
- Hayes, L.C.; Meers, A.; Tulley, K.; Sable, P.E.; Castagno, S.; Cilento, B.G. Interdisciplinary Collaboration in a Pediatric Urology Outpatient Clinic at a Tertiary Children’s Hospital: A Case Series. Urology 2022 , 169 , 191–195. [ Google Scholar ] [ CrossRef ]
- Getchell, K.; McCowan, K.; Whooley, E.; Dumais, C.; Rosenstock, A.; Cole, A.; DeGrazia, M. Child Life Specialists Decrease Procedure Time, Improve Experience, and Reduce Fear in an Outpatient Blood Drawing Lab (CLS Decrease Procedure Time). J. Patient Exp. 2022 , 9 , 23743735221105679. [ Google Scholar ] [ CrossRef ]
- Lloyd, J.K. Minimising stress for patients in the veterinary hospital: Why it is important and what can be done about it. Vet. Sci. 2017 , 4 , 22. [ Google Scholar ] [ CrossRef ]
- Riemer, S.; Heritier, C.; Windschnurer, I.; Pratsch, L.; Arhant, C.; Affenzeller, N. A Review on Mitigating Fear and Aggression in Dogs and Cats in a Veterinary Setting. Animals 2021 , 11 , 158. [ Google Scholar ] [ CrossRef ]
- Herron, M.E.; Shreyer, T. The pet-friendly veterinary practice: A guide for practitioners. Vet. Clin. Small Anim. Pract. 2014 , 44 , 451–481. [ Google Scholar ]
- Shedlock, D.J.; Silvestri, G.; Weiner, D.B. Monkeying around with HIV vaccines: Using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat. Rev. Immunol. 2009 , 9 , 717–728. [ Google Scholar ] [ CrossRef ]
- Phillips, K.A.; Bales, K.L.; Capitanio, J.P.; Conley, A.; Czoty, P.W.; ‘t Hart, B.A.; Hopkins, W.D.; Hu, S.L.; Miller, L.A.; Nader, M.A. Why primate models matter. Am. J. Primatol. 2014 , 76 , 801–827. [ Google Scholar ] [ CrossRef ]
- Heijmans, C.M.C.; de Groot, N.G.; Bontrop, R.E. Comparative genetics of the major histocompatibility complex in humans and nonhuman primates. Int. J. Immunogenet. 2020 , 47 , 243–260. [ Google Scholar ] [ CrossRef ]
- Veissier, I.; Boissy, A. Stress and welfare: Two complementary concepts that are intrinsically related to the animal’s point of view. Physiol. Behav. 2007 , 92 , 429–433. [ Google Scholar ] [ CrossRef ]
- Bloomsmith, M.A.; Perlman, J.E.; Hutchinson, E.; Sharpless, M. Behavioral management programs to promote laboratory animal welfare. In Management of Animal Care and Use Programs in Research, Education, and Testing ; CRC Press: Boca Raton, FL, USA, 2017; pp. 63–82. [ Google Scholar ]
- Broom, D.M.; Kirkden, R.D. Welfare, stress, behaviour and pathophysiology. In Veterinary Pathophysiology ; Blackwell: Ames, IA, USA, 2004; pp. 337–369. [ Google Scholar ]
- Moberg, G.P. Biological response to stress: Implications for animal welfare. In The Biology of Animal Stress: Basic Principles and Implications for Animal Welfare ; CABI Publishing: Wallingford, UK, 2000; pp. 1–21. [ Google Scholar ]
- Freeman, H.D.; Brosnan, S.F.; Hopper, L.M.; Lambeth, S.P.; Schapiro, S.J.; Gosling, S.D. Developing a comprehensive and comparative questionnaire for measuring personality in chimpanzees using a simultaneous top-down/bottom-up design. Am. J. Primatol. 2013 , 75 , 1042–1053. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Novak, M.A.; Hamel, A.F.; Kelly, B.J.; Dettmer, A.M.; Meyer, J.S. Stress, the HPA axis, and nonhuman primate well-being: A review. Appl. Anim. Behav. Sci. 2013 , 143 , 135–149. [ Google Scholar ] [ CrossRef ]
- Lefman, S.H.; Prittie, J.E. Psychogenic stress in hospitalized veterinary patients: Causation, implications, and therapies. J Vet Emerg. Crit. Care 2019 , 29 , 107–120. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Prescott, M.J.; Lidster, K. Improving quality of science through better animal welfare: The NC3Rs strategy. Lab Anim. 2017 , 46 , 152–156. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Graham, M.L.; Prescott, M.J. The multifactorial role of the 3Rs in shifting the harm-benefit analysis in animal models of disease. Eur. J. Pharmacol. 2015 , 759 , 19–29. [ Google Scholar ] [ CrossRef ]
- Schapiro, S.J. Effects of social manipulations and environmental enrichment on behavior and cell-mediated immune responses in rhesus macaques. Pharmacol. Biochem. Behav. 2002 , 73 , 271–278. [ Google Scholar ] [ CrossRef ]
- Schapiro, S.J.; Lambeth, S.P. Control, choice, and assessments of the value of behavioral management to nonhuman primates in captivity. J. Appl. Anim. Welf. Sci. 2007 , 10 , 39–47. [ Google Scholar ] [ CrossRef ]
- Schapiro, S.J.; Nehete, P.N.; Perlman, J.E.; Sastry, K.J. A comparison of cell-mediated immune responses in rhesus macaques housed singly, in pairs, or in groups. Appl. Anim. Behav. Sci. 2000 , 68 , 67–84. [ Google Scholar ] [ CrossRef ]
- Palmer, S.; Oppler, S.H.; Graham, M.L. Behavioral Management as a Coping Strategy for Managing Stressors in Primates: The Influence of Temperament and Species. Biology 2022 , 11 , 423. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Laule, G.E.; Bloomsmith, M.A.; Schapiro, S.J. The use of positive reinforcement training techniques to enhance the care, management, and welfare of primates in the laboratory. J. Appl. Anim. Welf. Sci. 2003 , 6 , 163–173. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Graham, M.L.; Rieke, E.F.; Mutch, L.A.; Zolondek, E.K.; Faig, A.W.; DuFour, T.A.; Munson, J.W.; Kittredge, J.A.; Schuurman, H.-J. Successful implementation of cooperative handling eliminates the need for restraint in a complex non-human primate disease model: Cooperative handling in a macaque disease model. J. Med. Primatol. 2012 , 41 , 89–106. [ Google Scholar ] [ CrossRef ]
- Graham, M.L.; Rieke, E.F.; Dunning, M.; Mutch, L.A.; Craig, A.M.; Zolondek, E.K.; Hering, B.J.; Schuurman, H.J.; Bianco, R.W. A novel alternative placement site and technique for totally implantable vascular access ports in non-human primates. J. Med. Primatol. 2009 , 38 , 204–212. [ Google Scholar ] [ CrossRef ]
- Plunkett, P.K.; Byrne, D.G.; Breslin, T.; Bennett, K.; Silke, B. Increasing wait times predict increasing mortality for emergency medical admissions. Eur. J. Emerg. Med. 2011 , 18 , 192–196. [ Google Scholar ] [ CrossRef ]
- Guttmann, A.; Schull, M.J.; Vermeulen, M.J.; Stukel, T.A. Association between waiting times and short term mortality and hospital admission after departure from emergency department: Population based cohort study from Ontario, Canada. Bmj 2011 , 342 , d2983. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Staley, M.; Conners, M.G.; Hall, K.; Miller, L.J. Linking stress and immunity: Immunoglobulin A as a non-invasive physiological biomarker in animal welfare studies. Horm. Behav. 2018 , 102 , 55–68. [ Google Scholar ] [ CrossRef ]
- Minton, K. Immunometabolism: Stress-induced macrophage polarization. Nat. Rev. Immunol. 2017 , 17 , 277. [ Google Scholar ] [ CrossRef ]
- Dhabhar, F.S. Enhancing versus suppressive effects of stress on immune function: Implications for immunoprotection and immunopathology. Neuroimmunomodulation 2009 , 16 , 300–317. [ Google Scholar ] [ CrossRef ]
- Black, P.H. Stress and the inflammatory response: A review of neurogenic inflammation. Brain Behav. Immun. 2002 , 16 , 622–653. [ Google Scholar ] [ CrossRef ]
- Marsland, A.L.; Walsh, C.; Lockwood, K.; John-Henderson, N.A. The effects of acute psychological stress on circulating and stimulated inflammatory markers: A systematic review and meta-analysis. Brain Behav. Immun. 2017 , 64 , 208–219. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Segerstrom, S.C.; Miller, G.E. Psychological stress and the human immune system: A meta-analytic study of 30 years of inquiry. Psychol. Bull. 2004 , 130 , 601–630. [ Google Scholar ] [ CrossRef ]
- Walburn, J.; Vedhara, K.; Hankins, M.; Rixon, L.; Weinman, J. Psychological stress and wound healing in humans: A systematic review and meta-analysis. J. Psychosom. Res. 2009 , 67 , 253–271. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Lacy, N.L.; Paulman, A.; Reuter, M.D.; Lovejoy, B. Why we don’t come: Patient perceptions on no-shows. Ann. Fam. Med. 2004 , 2 , 541–545. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Acar, D.; Güneş, Z. Factors affecting therapeutic compliance in patients with chronic renal failure: Anxiety, Depression, İllness Perception. Age 2018 , 61 , 14–19. [ Google Scholar ] [ CrossRef ]
- Volk, J.O.; Felsted, K.E.; Thomas, J.G.; Siren, C.W. Executive summary of the Bayer veterinary care usage study. J. Am. Vet. Med. Assoc. 2011 , 238 , 1275–1282. [ Google Scholar ] [ CrossRef ]
- Stellato, A. Assessing Strategies for Reducing Dog Fear during Routine Physical Examinations ; University of Guelph: Guelph, ON, Canada, 2019. [ Google Scholar ]
- Paré, W.P.; Glavin, G.B. Restraint stress in biomedical research: A review. Neurosci. Biobehav. Rev. 1986 , 10 , 339–370. [ Google Scholar ] [ CrossRef ]
- Shirasaki, Y.; Yoshioka, N.; Kanazawa, K.; Maekawa, T.; Horikawa, T.; Hayashi, T. Effect of physical restraint on glucose tolerance in cynomolgus monkeys. J. Med. Primatol. 2013 , 42 , 165–168. [ Google Scholar ] [ CrossRef ]
- Morakinyo, A.O.; Ajiboye, K.I.; Oludare, G.O.; Samuel, T.A. Restraint stress impairs glucose homeostasis through altered insulin signalling in Sprague-Dawley rat. Niger. J. Physiol. Sci. 2016 , 31 , 23–29. [ Google Scholar ]
- Zisberg, A.; Gur-Yaish, N. Older adults’ personal routine at time of hospitalization. Geriatr. Nurs. 2017 , 38 , 27–32. [ Google Scholar ] [ CrossRef ]
- Porock, D.; Clissett, P.; Harwood, R.H.; Gladman, J.R. Disruption, control and coping: Responses of and to the person with dementia in hospital. Ageing Soc. 2015 , 35 , 37–63. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Manemann, S.M.; Chamberlain, A.M.; Roger, V.L.; Griffin, J.M.; Boyd, C.M.; Cudjoe, T.K.; Jensen, D.; Weston, S.A.; Fabbri, M.; Jiang, R. Perceived social isolation and outcomes in patients with heart failure. J. Am. Heart Assoc. 2018 , 7 , e008069. [ Google Scholar ] [ CrossRef ]
- Barnes, T.L.; MacLeod, S.; Tkatch, R.; Ahuja, M.; Albright, L.; Schaeffer, J.A.; Yeh, C.S. Cumulative effect of loneliness and social isolation on health outcomes among older adults. Aging Ment. Health 2022 , 26 , 1327–1334. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Kohn, J.; Panagiotakopoulos, L.; Neigh, G.N. The effects of social experience on the stress system and immune function in nonhuman primates. In Social Inequalities in Health in Nonhuman Primates: The Biology of the Gradient ; Spring International: Eschlikon, Switzerland, 2016; pp. 49–77. [ Google Scholar ]
- Pahar, B.; Baker, K.C.; Jay, A.N.; Russell-Lodrigue, K.E.; Srivastav, S.K.; Aye, P.P.; Blanchard, J.L.; Bohm, R.P. Effects of social housing changes on immunity and vaccine-specific immune responses in adolescent male rhesus macaques. Front. Immunol. 2020 , 11 , 565746. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Hannibal, D.L.; Bliss-Moreau, E.; Vandeleest, J.; McCowan, B.; Capitanio, J. Laboratory rhesus macaque social housing and social changes: Implications for research. Am. J. Primatol. 2017 , 79 , e22528. [ Google Scholar ] [ CrossRef ]
- Pomerantz, O.; Baker, K.C.; Bellanca, R.U.; Bloomsmith, M.A.; Coleman, K.; Hutchinson, E.K.; Pierre, P.J.; Weed, J.L.; Consortium, N.P.R.C.B.M. Improving transparency—A call to include social housing information in biomedical research articles involving nonhuman primates. Am. J. Primatol. 2022 , 84 , e23378. [ Google Scholar ] [ CrossRef ]
- Green, S.M.; Rothrock, S.G.; Lynch, E.L.; Ho, M.; Harris, T.; Hestdalen, R.; Hopkins, G.A.; Garrett, W.; Westcott, K. Intramuscular ketamine for pediatric sedation in the emergency department: Safety profile in 1022 cases. Ann. Emerg. Med. 1998 , 31 , 688–697. [ Google Scholar ] [ CrossRef ]
- Lee, V.K.; Flynt, K.S.; Haag, L.M.; Taylor, D.K. Comparison of the effects of ketamine, ketamine-medetomidine, and ketamine-midazolam on physiologic parameters and anesthesia-induced stress in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. J. Am. Assoc. Lab Anim. Sci. 2010 , 49 , 57–63. [ Google Scholar ]
- van Haperen, M.; Preckel, B.; Eberl, S. Indications, contraindications, and safety aspects of procedural sedation. Curr. Opin. Anesthesiol. 2019 , 32 , 769–775. [ Google Scholar ] [ CrossRef ]
Click here to enlarge figure
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Oppler, S.H.; Palmer, S.D.; Phu, S.N.; Graham, M.L. The Role of Behavioral Management in Enhancing Clinical Care and Efficiency, Minimizing Social Disruption, and Promoting Welfare in Captive Primates. Vet. Sci. 2024 , 11 , 401. https://doi.org/10.3390/vetsci11090401
Oppler SH, Palmer SD, Phu SN, Graham ML. The Role of Behavioral Management in Enhancing Clinical Care and Efficiency, Minimizing Social Disruption, and Promoting Welfare in Captive Primates. Veterinary Sciences . 2024; 11(9):401. https://doi.org/10.3390/vetsci11090401
Oppler, Scott H., Sierra D. Palmer, Sydney N. Phu, and Melanie L. Graham. 2024. "The Role of Behavioral Management in Enhancing Clinical Care and Efficiency, Minimizing Social Disruption, and Promoting Welfare in Captive Primates" Veterinary Sciences 11, no. 9: 401. https://doi.org/10.3390/vetsci11090401
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals
AMA Journal of Ethics ®
Illuminating the art of medicine, nonhuman animal research.
Nonhuman animals have long been and continue to be routinely used in biomedical and behavioral research to promote human health. When SARS-CoV2 infections triggered a race to develop and scale global access to vaccines in 2019, two key innovations happened to the supply chain of nonhuman animals created, raised, and used for science: (1) experiments and trials regarded as essential were prioritized and (2) governments and researchers shortened vaccine production timelines. Clinical and public health urgency concentrated and acutely focused demand for live mammals—especially ferrets, guinea pigs, hamsters, mice, nonhuman primates, pigs, and rats—in ways that also intensified demand for efficient protocol designs and streamlined methods of human vaccine research and development. Reasonable people can still disagree about when, why, and how nonhuman animals should be sacrificed for human health, but we now know that human health endeavors, specifically vaccine development, flourish even when we sanction fewer nonhuman animals’ cultivation and deaths for science. This theme issue investigates what this revelation from the pandemic means from clinical, ethical, legal, and policy standpoints for the future of human-centered research.

Why Should We Care About What Using Nonhuman Animals in Human-Centered Research Suggests About Our Characters?

How Should Clinician-Researchers Model Regard for Nonhuman Animals Bred for and Used in Human-Centered Science?

According to Which Criteria Should We Determine Whether and When IACUCs Are Sufficient for Protecting the Welfare of Nonhuman Animals Used in Research?

Roles of Randomized Controlled Trials in Establishing Evidence-Based Gender-Affirming Care and Advancing Health Equity

How Might Corporations’ and Nonhuman Animals’ Personhood Compare Under the Fifth and Fourteenth Amendments?

Should Nonhuman Animals Be Recognized Legally as Persons?

With What Should We Replace Nonhuman Animals in Biomedical Research Protocols?

Which Concepts Are Key to Transitioning From Nonhuman Animal Models to Engineered Microphysiological Systems in Biomedical Research?

How Should Treatment of Animals Beyond the Lab Factor Into Institutional Review?

How Should the 3 R ’s Be Revised and Why?
The American Medical Association on the Ethics of Vivisection, 1880-1950

Does It Make Sense to Say Humans “Protect” Nonhuman Animals While Using Them to Promote Human Health Interests?
Nonhuman animal research has contributed to human health advancements but raises questions about the extent to which humans protect nonhuman animals during such endeavors. This series of drawings explores several ethics and empirical questions from a visual point of view.

Humanity and Inhumanity of Nonhuman Primate Research

Ethics Talk: Are the “3 R 's” Enough to Express Respect for Nonhuman Animals in Human-Centered Research?

Editorial Fellow Interview: “Why Should We Care About What Using Nonhuman Animals in Human-Centered Research Suggests About Our Characters?”

Author Interview: “How Might Corporations’ and Nonhuman Animals’ Personhood Compare Under the Fifth and Fourteenth Amendments?”

Author Interview: “Which Concepts Are Key to Transitioning From Nonhuman Animal Models to Engineered Microphysiological Systems in Biomedical Research?”

Author Interview: “With What Should We Replace Nonhuman Animals in Biomedical Research Protocols?”

Author Interview: “How Should Treatment of Animals Beyond the Lab Factor Into Institutional Review?”
Author Interview: “The American Medical Association on the Ethics of Vivisection, 1880-1950”
Author Interview: “Does It Make Sense to Say Humans ‘Protect’ Nonhuman Animals While Using Them to Promote Human Health Interests?”
Other issues, antimicrobial stewardship, harm reduction and opioid use disorder, standards in medical-legal partnerships.
- All subject areas
- Agricultural and Biological Sciences
- Arts and Humanities
- Biochemistry, Genetics and Molecular Biology
- Business, Management and Accounting
- Chemical Engineering
- Computer Science
- Decision Sciences
- Earth and Planetary Sciences
- Economics, Econometrics and Finance
- Engineering
- Environmental Science
- Health Professions
- Immunology and Microbiology
- Materials Science
- Mathematics
- Multidisciplinary
- Neuroscience
- Pharmacology, Toxicology and Pharmaceutics
- Physics and Astronomy
- Social Sciences
- All subject categories
- Acoustics and Ultrasonics
- Advanced and Specialized Nursing
- Aerospace Engineering
- Agricultural and Biological Sciences (miscellaneous)
- Agronomy and Crop Science
- Algebra and Number Theory
- Analytical Chemistry
- Anesthesiology and Pain Medicine
- Animal Science and Zoology
- Anthropology
- Applied Mathematics
- Applied Microbiology and Biotechnology
- Applied Psychology
- Aquatic Science
- Archeology (arts and humanities)
- Architecture
- Artificial Intelligence
- Arts and Humanities (miscellaneous)
- Assessment and Diagnosis
- Astronomy and Astrophysics
- Atmospheric Science
- Atomic and Molecular Physics, and Optics
- Automotive Engineering
- Behavioral Neuroscience
- Biochemistry
- Biochemistry, Genetics and Molecular Biology (miscellaneous)
- Biochemistry (medical)
- Bioengineering
- Biological Psychiatry
- Biomaterials
- Biomedical Engineering
- Biotechnology
- Building and Construction
- Business and International Management
- Business, Management and Accounting (miscellaneous)
- Cancer Research
- Cardiology and Cardiovascular Medicine
- Care Planning
- Cell Biology
- Cellular and Molecular Neuroscience
- Ceramics and Composites
- Chemical Engineering (miscellaneous)
- Chemical Health and Safety
- Chemistry (miscellaneous)
- Chiropractics
- Civil and Structural Engineering
- Clinical Biochemistry
- Clinical Psychology
- Cognitive Neuroscience
- Colloid and Surface Chemistry
- Communication
- Community and Home Care
- Complementary and Alternative Medicine
- Complementary and Manual Therapy
- Computational Mathematics
- Computational Mechanics
- Computational Theory and Mathematics
- Computer Graphics and Computer-Aided Design
- Computer Networks and Communications
- Computer Science Applications
- Computer Science (miscellaneous)
- Computer Vision and Pattern Recognition
- Computers in Earth Sciences
- Condensed Matter Physics
- Conservation
- Control and Optimization
- Control and Systems Engineering
- Critical Care and Intensive Care Medicine
- Critical Care Nursing
- Cultural Studies
- Decision Sciences (miscellaneous)
- Dental Assisting
- Dental Hygiene
- Dentistry (miscellaneous)
- Dermatology
- Development
- Developmental and Educational Psychology
- Developmental Biology
- Developmental Neuroscience
- Discrete Mathematics and Combinatorics
- Drug Discovery
- Drug Guides
- Earth and Planetary Sciences (miscellaneous)
- Earth-Surface Processes
- Ecological Modeling
- Ecology, Evolution, Behavior and Systematics
- Economic Geology
- Economics and Econometrics
- Economics, Econometrics and Finance (miscellaneous)
- Electrical and Electronic Engineering
- Electrochemistry
- Electronic, Optical and Magnetic Materials
- Emergency Medical Services
- Emergency Medicine
- Emergency Nursing
- Endocrine and Autonomic Systems
- Endocrinology
- Endocrinology, Diabetes and Metabolism
- Energy Engineering and Power Technology
- Energy (miscellaneous)
- Engineering (miscellaneous)
- Environmental Chemistry
- Environmental Engineering
- Environmental Science (miscellaneous)
- Epidemiology
- Experimental and Cognitive Psychology
- Family Practice
- Filtration and Separation
- Fluid Flow and Transfer Processes
- Food Animals
- Food Science
- Fuel Technology
- Fundamentals and Skills
- Gastroenterology
- Gender Studies
- Genetics (clinical)
- Geochemistry and Petrology
- Geography, Planning and Development
- Geometry and Topology
- Geotechnical Engineering and Engineering Geology
- Geriatrics and Gerontology
- Gerontology
- Global and Planetary Change
- Hardware and Architecture
- Health Informatics
- Health Information Management
- Health Policy
- Health Professions (miscellaneous)
- Health (social science)
- Health, Toxicology and Mutagenesis
- History and Philosophy of Science
- Horticulture
- Human Factors and Ergonomics
- Human-Computer Interaction
- Immunology and Allergy
- Immunology and Microbiology (miscellaneous)
- Industrial and Manufacturing Engineering
- Industrial Relations
- Infectious Diseases
- Information Systems
- Information Systems and Management
- Inorganic Chemistry
- Insect Science
- Instrumentation
- Internal Medicine
- Issues, Ethics and Legal Aspects
- Leadership and Management
- Library and Information Sciences
- Life-span and Life-course Studies
- Linguistics and Language
- Literature and Literary Theory
- LPN and LVN
- Management Information Systems
- Management, Monitoring, Policy and Law
- Management of Technology and Innovation
- Management Science and Operations Research
- Materials Chemistry
- Materials Science (miscellaneous)
- Maternity and Midwifery
- Mathematical Physics
- Mathematics (miscellaneous)
- Mechanical Engineering
- Mechanics of Materials
- Media Technology
- Medical and Surgical Nursing
- Medical Assisting and Transcription
- Medical Laboratory Technology
- Medical Terminology
- Medicine (miscellaneous)
- Metals and Alloys
- Microbiology
- Microbiology (medical)
- Modeling and Simulation
- Molecular Biology
- Molecular Medicine
- Nanoscience and Nanotechnology
- Nature and Landscape Conservation
- Neurology (clinical)
- Neuropsychology and Physiological Psychology
- Neuroscience (miscellaneous)
- Nuclear and High Energy Physics
- Nuclear Energy and Engineering
- Numerical Analysis
- Nurse Assisting
- Nursing (miscellaneous)
- Nutrition and Dietetics
- Obstetrics and Gynecology
- Occupational Therapy
- Ocean Engineering
- Oceanography
- Oncology (nursing)
- Ophthalmology
- Oral Surgery
- Organic Chemistry
- Organizational Behavior and Human Resource Management
- Orthodontics
- Orthopedics and Sports Medicine
- Otorhinolaryngology
- Paleontology
- Parasitology
- Pathology and Forensic Medicine
- Pathophysiology
- Pediatrics, Perinatology and Child Health
- Periodontics
- Pharmaceutical Science
- Pharmacology
- Pharmacology (medical)
- Pharmacology (nursing)
- Pharmacology, Toxicology and Pharmaceutics (miscellaneous)
- Physical and Theoretical Chemistry
- Physical Therapy, Sports Therapy and Rehabilitation
- Physics and Astronomy (miscellaneous)
- Physiology (medical)
- Plant Science
- Political Science and International Relations
- Polymers and Plastics
- Process Chemistry and Technology
- Psychiatry and Mental Health
- Psychology (miscellaneous)
- Public Administration
- Public Health, Environmental and Occupational Health
- Pulmonary and Respiratory Medicine
- Radiological and Ultrasound Technology
- Radiology, Nuclear Medicine and Imaging
- Rehabilitation
- Religious Studies
- Renewable Energy, Sustainability and the Environment
- Reproductive Medicine
- Research and Theory
- Respiratory Care
- Review and Exam Preparation
- Reviews and References (medical)
- Rheumatology
- Safety Research
- Safety, Risk, Reliability and Quality
- Sensory Systems
- Signal Processing
- Small Animals
- Social Psychology
- Social Sciences (miscellaneous)
- Social Work
- Sociology and Political Science
- Soil Science
- Space and Planetary Science
- Spectroscopy
- Speech and Hearing
- Sports Science
- Statistical and Nonlinear Physics
- Statistics and Probability
- Statistics, Probability and Uncertainty
- Strategy and Management
- Stratigraphy
- Structural Biology
- Surfaces and Interfaces
- Surfaces, Coatings and Films
- Theoretical Computer Science
- Tourism, Leisure and Hospitality Management
- Transplantation
- Transportation
- Urban Studies
- Veterinary (miscellaneous)
- Visual Arts and Performing Arts
- Waste Management and Disposal
- Water Science and Technology
- All regions / countries
- Asiatic Region
- Eastern Europe
- Latin America
- Middle East
- Northern America
- Pacific Region
- Western Europe
- ARAB COUNTRIES
- IBEROAMERICA
- NORDIC COUNTRIES
- Afghanistan
- Bosnia and Herzegovina
- Brunei Darussalam
- Czech Republic
- Dominican Republic
- Netherlands
- New Caledonia
- New Zealand
- Papua New Guinea
- Philippines
- Puerto Rico
- Russian Federation
- Saudi Arabia
- South Africa
- South Korea
- Switzerland
- Syrian Arab Republic
- Trinidad and Tobago
- United Arab Emirates
- United Kingdom
- United States
- Vatican City State
- Book Series
- Conferences and Proceedings
- Trade Journals

- Citable Docs. (3years)
- Total Cites (3years)

| Title | Type | --> | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | journal | 4.891 Q1 | 57 | 223 | 615 | 5851 | 6699 | 317 | 10.33 | 26.24 | 47.92 | ||
| 2 | journal | 2.665 Q1 | 60 | 16 | 63 | 1926 | 640 | 60 | 8.82 | 120.38 | 38.64 | ||
| 3 | journal | 1.801 Q1 | 177 | 194 | 675 | 14977 | 2881 | 669 | 3.39 | 77.20 | 34.14 | ||
| 4 | journal | 1.744 Q1 | 212 | 400 | 1064 | 29436 | 7573 | 1062 | 6.59 | 73.59 | 37.28 | ||
| 5 | journal | 1.735 Q1 | 168 | 862 | 2056 | 40643 | 21572 | 2056 | 9.27 | 47.15 | 30.69 | ||
| 6 | journal | 1.735 Q1 | 90 | 20 | 121 | 1846 | 671 | 116 | 5.40 | 92.30 | 29.24 | ||
| 7 | journal | 1.585 Q1 | 134 | 174 | 604 | 13239 | 4509 | 597 | 7.28 | 76.09 | 36.18 | ||
| 8 | journal | 1.563 Q1 | 49 | 21 | 95 | 1152 | 790 | 95 | 3.09 | 54.86 | 29.17 | ||
| 9 | journal | 1.453 Q1 | 71 | 157 | 379 | 11196 | 2567 | 378 | 6.51 | 71.31 | 40.83 | ||
| 10 | journal | 1.319 Q1 | 53 | 148 | 351 | 10465 | 2411 | 351 | 6.59 | 70.71 | 37.93 | ||
| 11 | journal | 1.302 Q1 | 55 | 74 | 146 | 3638 | 1702 | 146 | 9.77 | 49.16 | 28.06 | ||
| 12 | journal | 1.286 Q1 | 32 | 111 | 296 | 6480 | 1099 | 173 | 3.64 | 58.38 | 35.89 | ||
| 13 | journal | 1.219 Q1 | 229 | 709 | 2758 | 38967 | 11638 | 2747 | 3.93 | 54.96 | 43.83 | ||
| 14 | journal | 1.197 Q1 | 17 | 22 | 77 | 2101 | 212 | 75 | 2.77 | 95.50 | 29.93 | ||
| 15 | journal | 1.159 Q1 | 36 | 87 | 224 | 7462 | 975 | 222 | 3.92 | 85.77 | 36.14 | ||
| 16 | journal | 1.142 Q1 | 72 | 45 | 154 | 3621 | 417 | 154 | 2.45 | 80.47 | 30.61 | ||
| 17 | journal | 1.126 Q1 | 22 | 66 | 198 | 5670 | 958 | 196 | 4.86 | 85.91 | 45.98 | ||
| 18 | journal | 1.082 Q1 | 8 | 45 | 41 | 3348 | 236 | 40 | 5.76 | 74.40 | 36.60 | ||
| 19 | journal | 1.080 Q1 | 169 | 916 | 2199 | 43606 | 10264 | 2182 | 4.39 | 47.60 | 43.09 | ||
| 20 | journal | 1.059 Q1 | 19 | 18 | 59 | 1411 | 240 | 56 | 3.62 | 78.39 | 36.75 | ||
| 21 | journal | 1.040 Q1 | 132 | 81 | 447 | 5455 | 987 | 405 | 2.21 | 67.35 | 40.00 | ||
| 22 | journal | 1.026 Q1 | 87 | 14 | 149 | 1408 | 395 | 147 | 2.69 | 100.57 | 41.04 | ||
| 23 | book series | 1.026 Q1 | 41 | 4 | 12 | 696 | 38 | 12 | 2.00 | 174.00 | 28.57 | ||
| 24 | journal | 1.025 Q1 | 87 | 93 | 242 | 4941 | 958 | 241 | 3.53 | 53.13 | 38.64 | ||
| 25 | journal | 1.017 Q1 | 211 | 497 | 1776 | 25948 | 3655 | 1341 | 1.98 | 52.21 | 39.63 | ||
| 26 | journal | 1.010 Q1 | 122 | 296 | 923 | 15572 | 3946 | 907 | 4.29 | 52.61 | 45.49 | ||
| 27 | journal | 1.000 Q1 | 132 | 188 | 694 | 8572 | 4136 | 692 | 4.99 | 45.60 | 27.97 | ||
| 28 | journal | 0.993 Q1 | 95 | 112 | 476 | 9570 | 1312 | 475 | 3.02 | 85.45 | 27.12 | ||
| 29 | journal | 0.979 Q1 | 48 | 18 | 83 | 909 | 554 | 83 | 3.41 | 50.50 | 42.42 | ||
| 30 | journal | 0.949 Q1 | 81 | 295 | 858 | 15446 | 4240 | 842 | 5.09 | 52.36 | 37.84 | ||
| 31 | journal | 0.941 Q1 | 121 | 202 | 569 | 13058 | 2714 | 569 | 4.52 | 64.64 | 44.80 | ||
| 32 | journal | 0.932 Q1 | 14 | 20 | 131 | 1393 | 534 | 127 | 4.20 | 69.65 | 36.36 | ||
| 33 | journal | 0.924 Q1 | 189 | 183 | 710 | 13778 | 1640 | 694 | 2.19 | 75.29 | 44.33 | ||
| 34 | journal | 0.919 Q1 | 11 | 44 | 92 | 2309 | 283 | 91 | 3.08 | 52.48 | 44.37 | ||
| 35 | journal | 0.917 Q1 | 73 | 40 | 127 | 2850 | 355 | 127 | 2.79 | 71.25 | 41.67 | ||
| 36 | journal | 0.901 Q1 | 35 | 40 | 87 | 2115 | 273 | 87 | 3.33 | 52.88 | 46.15 | ||
| 37 | journal | 0.889 Q1 | 143 | 515 | 1335 | 22708 | 4208 | 1293 | 3.26 | 44.09 | 52.03 | ||
| 38 | journal | 0.881 Q1 | 9 | 57 | 75 | 3227 | 258 | 73 | 3.44 | 56.61 | 38.69 | ||
| 39 | journal | 0.868 Q1 | 139 | 128 | 466 | 10385 | 1083 | 458 | 2.13 | 81.13 | 46.62 | ||
| 40 | journal | 0.867 Q1 | 68 | 5 | 56 | 507 | 215 | 55 | 4.12 | 101.40 | 38.46 | ||
| 41 | journal | 0.856 Q1 | 44 | 124 | 217 | 8304 | 730 | 203 | 3.57 | 66.97 | 36.55 | ||
| 42 | journal | 0.855 Q1 | 20 | 41 | 120 | 3191 | 266 | 120 | 2.45 | 77.83 | 18.65 | ||
| 43 | journal | 0.853 Q1 | 55 | 94 | 353 | 5995 | 1144 | 256 | 3.13 | 63.78 | 41.51 | ||
| 44 | journal | 0.843 Q1 | 53 | 108 | 140 | 6434 | 559 | 138 | 4.08 | 59.57 | 38.25 | ||
| 45 | journal | 0.832 Q1 | 90 | 145 | 328 | 10724 | 733 | 314 | 2.26 | 73.96 | 33.61 | ||
| 46 | journal | 0.823 Q1 | 42 | 446 | 1163 | 28308 | 4584 | 1163 | 3.64 | 63.47 | 38.34 | ||
| 47 | journal | 0.818 Q1 | 25 | 90 | 249 | 5725 | 652 | 240 | 2.40 | 63.61 | 40.63 | ||
| 48 | journal | 0.812 Q1 | 75 | 53 | 207 | 2629 | 397 | 198 | 1.38 | 49.60 | 31.25 | ||
| 49 | journal | 0.803 Q1 | 135 | 140 | 478 | 10621 | 998 | 469 | 1.77 | 75.86 | 43.55 | ||
| 50 | journal | 0.788 Q1 | 88 | 48 | 141 | 4788 | 561 | 129 | 3.93 | 99.75 | 30.64 |

Follow us on @ScimagoJR Scimago Lab , Copyright 2007-2024. Data Source: Scopus®

Cookie settings
Cookie Policy
Legal Notice
Privacy Policy
ORIGINAL RESEARCH article
Development and validation of a machine learning model for clinical wellness visit classification in cats and dogs.

- IDEXX Laboratories, Inc., Westbrook, ME, United States
Introduction: Early disease detection in veterinary care relies on identifying subclinical abnormalities in asymptomatic animals during wellness visits. This study introduces a model designed to distinguish between wellness and other types of veterinary visits.
Objectives: The purpose of this study is to validate the use of a visit classification model compared to manual classification of veterinary visits by three board-certified veterinarians.
Materials and methods: The algorithm was initially trained using a Gradient Boosting Machine model with a dataset of 11,105 clinical visits from 2012 to 2017 involving 655 animals (85.3% dogs and 14.7% cats) across 544 U.S. veterinary practices. Three validators were tasked with classifying 400 visits, including both wellness and other types of visits, selected randomly from the same database used for initial model training, aiming to maintain consistency and relevance between the training and application phases; visit classifications were subsequently categorized into “wellness” or “other” based on majority consensus among validators to assess the model’s performance in identifying wellness visits.
Results: The model demonstrated a specificity of 0.94 (95% CI: 0.91 to 0.96), implying its accuracy in distinguishing non-wellness visits. The model had a sensitivity of 0.86 (95% CI: 0.80 to 0.92), indicating its ability to correctly identify wellness visits as compared to the annotations provided by veterinary experts. The balanced accuracy, calculated as 0.90 (95% CI: 0.87 to 0.93), further confirms the model’s overall effectiveness.
Clinical significance: The model exhibits high specificity and sensitivity, ensuring accurate identification of a high proportion of wellness visits. Overall, this model holds promise for advancing research on preventive care’s role in subclinical disease identification, but prospective studies are needed for validation.
Introduction
In recent years, artificial intelligence (AI) and machine learning (ML) models have begun to transform veterinary medicine, enhancing diagnostic capabilities and treatment methodologies ( 1 ). These technologies are increasingly used to analyze complex data and improve the accuracy of diagnoses across a variety of conditions. For example, machine learning applications in veterinary medicine have shown significant potential in areas ranging from remote sensing in wildlife monitoring to advanced imaging techniques for companion animals ( 2 , 3 ). Such advancements underscore the growing importance of AI in the field, setting the stage for more specialized applications.
Medical records can provide a wealth of information to support studies of disease prevalence. Within the human medical field, standardized medical coding for insurance and the International Coding of Disease (ICD-11) can facilitate classification of visit types or diagnoses across clinics and hospitals ( 4 ). Veterinary medicine lacks a similar classification system that allows for standardized recognition of clinical behavior at visits, and this has been a barrier to studies investigating clinical behavior, results, or outcomes in the field ( 5 ). Machine learning techniques are increasingly employed to structure and derive insights from the vast amount of structured, semi-structured, and unstructured data in veterinary medicine, facilitating automated extraction of valuable information from clinical narratives and improving both animal and human health outcomes ( 1 , 6 – 8 ).
Wellness or preventive care visits play an important role in the veterinary-client-patient relationship and provide an opportunity to educate clients and identify subclinical disease ( 9 , 10 ). While companion animal preventative health guidelines to exist, there can be substantial variation between clinics in what is included within a wellness visit and there is no general agreement on what defines a wellness visit or what should be included in wellness care ( 9 – 13 ). Depending on the clinic, a wellness visit could be defined only as the examination of apparently healthy animals with no health concerns, to visits that include preventive care and routine laboratory testing of blood, feces and urine in addition to the physical exam. There is limited information on the results of routine bloodwork—meaning tests ordered without intending diagnosis or monitoring—at various life stages. Most studies that have explored laboratory results in healthy dogs and cats typically have a narrow scope ( 14 – 17 ). They might concentrate on one specific breed, consider only a restricted range of analytes, focus primarily on older pets, or implement a strict definition of what defines “healthy” pets. Currently, large scale real-world evidence (RWE) studies on the value of wellness visits are limited. A major barrier in the collection of large-scale RWE studies is the inability to determine if a clinical examination constitutes a wellness visit due to the non-uniform collection of clinical information.
This study aims to validate a machine learning model designed to classify wellness visits for dogs and cats that have been presented to veterinary practices across the United States. The performance of the model will be benchmarked as compared to the consensus results of three licensed veterinarians who classified a clinical examination as a wellness visit or other visit.
Model training
The purpose of the visit classification model was to use electronic practice information management records to determine why a pet owner visited their veterinarian (i.e., Why did the pet owner bring their animal to the practice?). A patient visit was defined as when a patient (or patient’s owner) comes to the clinic and obtains one or more products or services on behalf of an animal. The visit begins when the animal walks through the clinic door and ends when the animal leaves the clinic. A patient visit may also be when an animal owner comes to the clinic and obtains some product on behalf of the animal. Each visit was reviewed and classified based on what the intent of the visit was by the pet owner.
Several factors were used to ascertain the intention of a visit including the time since last visit, appointment notes, invoice items, and any available medical notes. The primary sources of information were the medical notes or the stated reasons for the visit. In cases where medical notes were unavailable, transaction details were evaluated. Entries explicitly mentioning ‘yearly wellness exam’ or ‘6-month exam’ were classified as wellness visits. Additionally, if a list of services included routine procedures such as vaccinations, ear swabs, fecal tests, or routine blood work fitting the typical timing of a yearly or six-month checkup, the visit was categorized as wellness. However, wellness visits that coincided with grooming or boarding services were not classified as wellness, since the primary intent was deemed to be grooming or boarding, with medical services provided as a convenience. Visits with unclear intent based on the provided information, or those recorded as mere administrative line items not representing actual activity for the pet, were excluded from our analysis.
During the initial human annotation process, visits were first classified into clinical or non-clinical visit categories. Visits were then classified further as wellness, non-wellness, and non-clinical visits (consisting of boarding, grooming, and retail). Annotators were masked to visit classifications applied by other annotators. Visits were organized by patient at a single clinic. A custom annotation tool was developed to anonymously monitor annotator consensus. It tracked individual annotator metrics, training set agreement, and group agreement on new labels (blind to individual annotators).
A selection of 11,105 clinical visits from 2012 to 2017 was used to train our model. These visits involved 655 animals (85.3% dogs and 14.7% cats) from 544 veterinary establishments in the United States. These visits were randomly selected from the database using the default random number generator (RNG) in base R, the Mersenne-Twister algorithm, to ensure that each visit had an equal chance of being chosen ( 18 ). The median duration of a visit was one day (IQR: 1.0–1.0 days) and the median number of transaction line items per visit was two transactions (IQR: 1–5 transactions). We classified the visits into four categories: wellness (24.5%), non-wellness (23.1%), non-clinical (42.5%), and unknown (10.0%). A single visit could have multiple labels with the exception of a non-clinical visit which was defined to be mutually exclusive to all the other categories.
During the model development, to annotate the visits for intent, two methods were used. First, a pair of veterinarians to label each of the 5,984 visits in the preliminary phase ( Supplementary Figure S1A ). They agreed on 5,058 visits and labeled the remaining 926 as unknown. These unknown visits were excluded from the initial model training. Second, one of the six board-certified veterinarians labeled an additional 5,121 visits. These visits were added to the training dataset along with the agreed-upon 5,058 visits from the preliminary phase.
A comprehensive set of features was used to classify the visits into one of the four categories: wellness, non-wellness, non-clinical and unknown using a Gradient Boosting Machine (GBM) model using the H20 3.20.0.2 and R version 3.5.1 ( 19 , 20 ). GBM is a decision tree-based algorithm that creates a single model by adding together output from many small, weak decision trees. Each tree is constructed in sequence to fix the errors made by all the previous trees. For a binary classifier (one with 2 classes), the output of the model is the probability that the data provided is in the class (or not in the class). GBM chosen over other methods due to its superior predictive performance and robustness in handling complex non-linear relationships within the data. Furthermore, its ability to provide insights into feature importance and manage diverse data types makes it particularly suitable for our veterinary visit classification task ( 21 ). A simple grid search approach was employed to optimize the performance of the Gradient Boosting Machine (GBM). To ensure the validity of the results, the data used for hyperparameter selection did not overlap with any validation or test datasets, thereby avoiding data leakage and ensuring that performance metrics accurately reflect the model’s generalization capabilities. The features used to classify a visit encompassed various demographic and clinical aspects of the visits, including species, age, the number of items on the invoice, total visit cost, days since the last visit, days since the second to last visit, days since the third to last visit, transaction labels, vaccine terms, prescriptions, medical notes (examination type, procedure, etc.), appointment notes words, and appointment reason for visit words. The feature set included all the features available to the veterinarians at the time of annotation. By leveraging this diverse array of features, the model aimed to capture nuanced patterns and relationships in the data, ultimately enhancing its ability to distinguish between wellness visits and other visits in veterinary clinics. The classification model for the classification of wellness visits, was trained employing a 5-fold cross-validation approach, with the F1 score used to select the final trained model. The final GBM model was composed of 111 trees with a maximum depth of 7. The most influential features include vaccine terms, prescriptions, vector born disease test, and visit total cost. Other notable features are laboratory tests medical service, various time-based metrics such as days since the last visit and visit interval, animal age, and several other visit-related metrics. This process resulted in performance metrics including an F1 score of 0.93, a recall of 0.93, a precision of 0.93, and a specificity of 0.97 for the detection of wellness visits as measured on a held-out test dataset of 1,003 visits that was randomly selected from the training set. The held-out set was separate from the training set.
Training validation annotators
The three veterinarian validators were put through an education period using 125 visits (25 visits from wellness, non-wellness and 75 from the non-clinical visit category) to become familiar with the annotation tool ( Supplementary Figure S1B ). These visits were selected from the initial 5,058 visits where the label was agreed upon by two veterinarians and were used to train the model (see Model Training Section above). The veterinarians were granted access to the labels that had been collaboratively determined by the two initial veterinarians who categorized the visit. As part of the education process, round table virtual discussions were allowed to gain alignment on classification of visits. These discussions were facilitated by an expert veterinarian involved in the initial training of the model (MC) and the data scientists involved in the development of the model (JR, DM).
After the education period, each of the validators were assessed for agreement to a random selection of 100 visits that matched the distribution of visit types seen in production. The veterinarians were masked to the label of the visits for this assessment. These visits were selected from the initial 5,058 visits where the label was agreed upon by two veterinarians and were used to train the model (see Model Training Section above). A benchmark of 85% agreement (defined pre-hoc ) to the initial agreed upon label by the two veterinarians used to develop the training dataset was required for the validators to continue to the validation study.
Development of validation dataset
After the educational period, the three validators were assigned the task of classifying the same 400 visits, distinguishing between wellness visits and other types of visits. To ensure that these 400 visits reflected the distribution of the population of visits expected in a live environment, they were randomly selected from the same database that was used for the initial training of the model, but were not part of the data used to train the model. This was done to ensure consistency and relevance between the training and application phases. To assess the performance of the model for the identification of wellness visits, the results were split into two categories: (1) “wellness” containing the wellness visit type; (2) “other” containing all visits not defined as a wellness visit (for example non-wellness and non-clinical visits). The reference label for each visit was then generated by using the majority consensus for “wellness” or “other” by each of the validators.
Statistical analysis
A sample size of 400 visits was considered adequate for the identification of wellness visits based off bootstrap simulation using one of the holdout datasets from the k-fold cross validation from the training dataset. A total of 800 visits (double the required amount based on sample size calculation) were collected. In the case that the validators finished their caseload with additional time, they were allowed to continue annotating visits until budget ran out. Consequently, the final validation dataset comprised 622 visits. Initial assessment of validator performance against each other was compared using percent exact match across all three raters and Fleiss Kappa for agreement. The performance of the model in accurately classifying wellness and other visits was evaluated using several metrics, including Sensitivity (Recall), Specificity, Positive Predictive Value (PPV, Precision), Negative Predictive Value (NPV), F1 Score, Balanced Accuracy, Matthews Correlation Coefficient, and Jaccard Index. To address the absence of a gold standard, sensitivity, specificity, and prevalence were estimated using the Expectation–Maximization (EM) algorithm with conditional independence, providing additional measures of the model’s performance ( 22 , 23 ). To determine the confidence intervals and estimates for these metrics, a bootstrapping approach was applied. Specifically, 2,000 bootstrap samples were generated by randomly sampling visits with the replacement from the original dataset. Bootstrap validation and calibration plots were performed using the Hmisc and rms packages ( 24 , 25 ). Statistical analysis was done using R version 4.0.2 and various helper functions from the tidyverse ( 19 , 26 ). Data visualization was generated using ggplot2 ( 27 ).
A total of 622 total visits were classified by all three trained validating veterinarians. Most were dog visits (78.2%, n = 487) with 135 cat visits (21.7%). The median age of dogs was 6.0 years (IQR: 2.0–9.3 years) and of cats was 6.0 years (1.7–12.0) with a single cat with no recorded age. A full list of demographic information can be found in Table 1 .
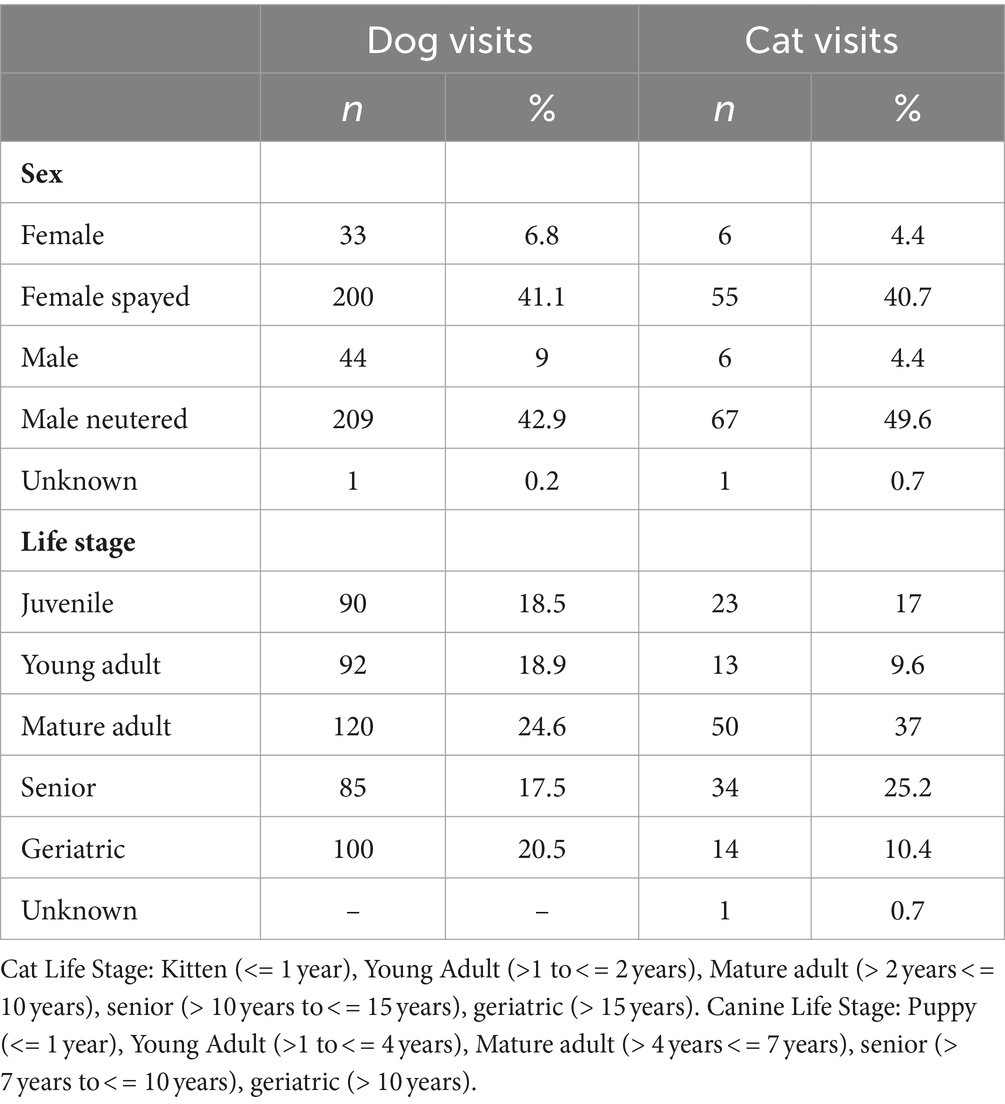
Table 1 . Pet demographic information from 622 veterinary visits.
Interobserver agreement
Of the 622 visits, all three trained validators agreed on the classification of 96.9% (602) of the visits with 457 (73.5%) visits being classified as other and 146 (23.5%) being classified as wellness (Fleiss Kappa = 0.91, Supplementary Table S1 ; Appendix S1 ). The proportion of visits that have a reference label of wellness is 23.3% ( n = 145), with 24.8% ( n = 121) wellness visits classified for dogs and 17.8% ( n = 24) classified as wellness for cats.
Model performance
The model demonstrated a specificity of 0.94 (95% CI: 0.91 to 0.96), implying its accuracy in distinguishing non-wellness visits ( Tables 2 , 3 ). The model had a sensitivity of 0.86 (95% CI: 0.80 to 0.92), indicating its ability to correctly identify wellness visits as compared to the annotations provided by veterinary experts ( Tables 2 , 3 ). This suggests that the model accurately identified 86% of wellness visits. These results indicate that the model accurately recognized 94% of non-wellness and had a low false positive rate. The balanced accuracy, calculated as 0.90 (95% CI: 0.87 to 0.93), further confirms the model’s overall effectiveness ( Table 3 ). In addition, the model was observed to be well calibrated across the predictive range ( Supplementary Figure S2 ). Employing the conditional independence model with the EM algorithm to account for an imperfect gold standard, we obtained a specificity of 0.97 (95% CI: 0.95–0.99) and a sensitivity of 0.85 (95% CI: 0.79–0.90). Additional measures of model performance are found in Table 3 and in Appendix 2 .
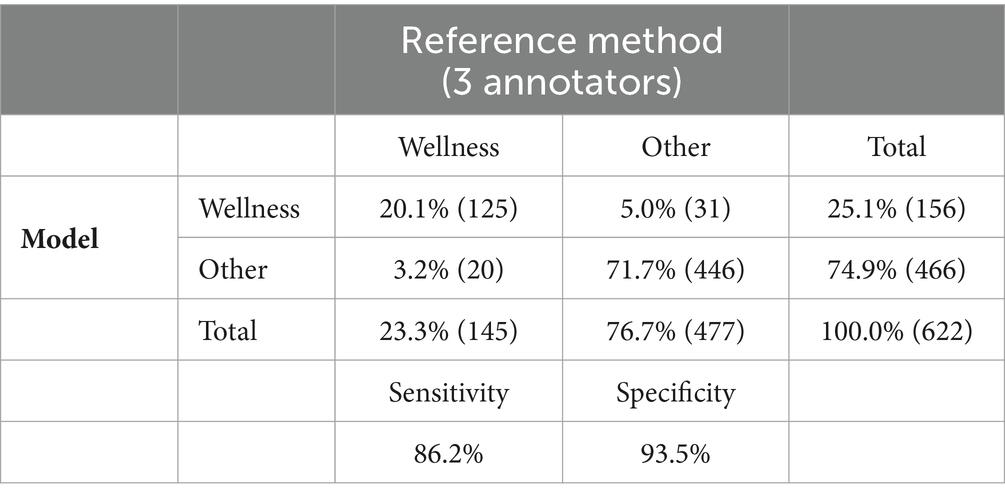
Table 2 . Contingency table of model performance to correctly identify wellness visits as compared to reference method (majority of three veterinarians).
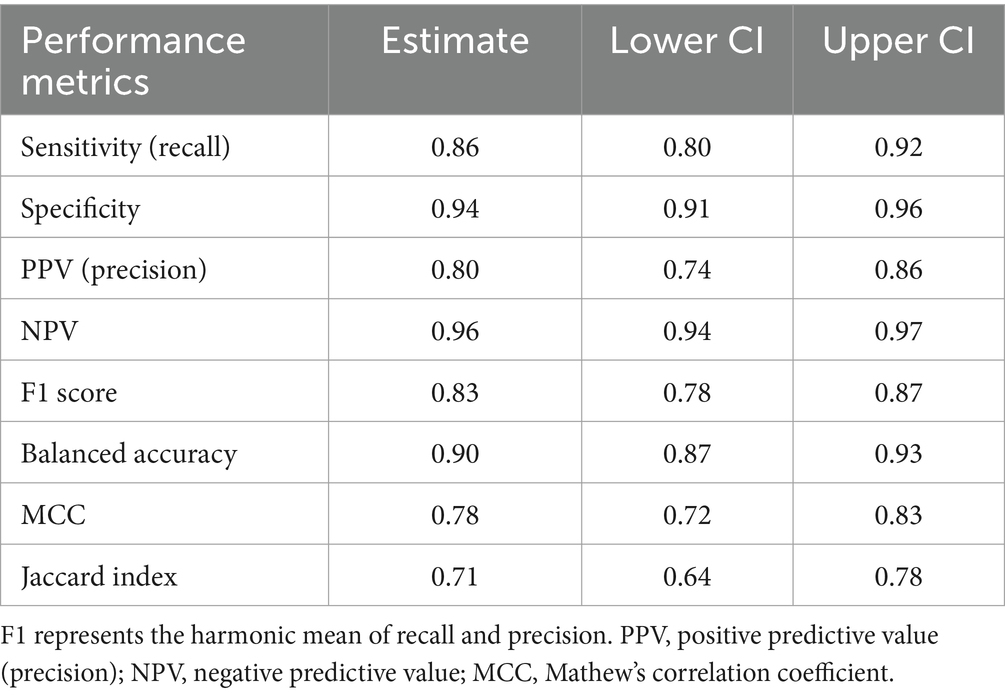
Table 3 . Bootstrapped model performance of model and 95% confidence interval estimates.
Early disease detection in veterinary care depends on the identification of subclinical abnormalities in asymptomatic animals which could be evaluated during a wellness visit ( 14 ). Providing a method to determine the type of veterinary visit is essential to determining the benefit of wellness visits. The model developed for this study demonstrated strong specificity and sensitivity, suggesting a robust ability to distinguish between wellness and other visits. The high specificity and sensitivity confirm that the model was able to accurately identify a high proportion of wellness visits. A high specificity in our classification model minimizes the risks associated with incorrectly categorizing other visits as wellness visits. Such a misclassification would have more serious consequences compared to inadvertently classifying a wellness visit as a non-wellness one, as it could potentially lead to overlooking individuals who are in actual need of medical attention or intervention.
This study has limitations that warrant consideration. Primarily, the identification and classification of visit types were conducted using data from specific practice information management systems, which may limit the model’s generalizability across different clinical settings and necessitates additional validation. Furthermore, evaluating visit types is not commonly integrated into the standard veterinary clinical workflow. Therefore, direct comparisons of the model’s capability to accurately identify wellness visits with those documented by a single veterinarian may prove difficult to interpret. To address this, we propose the collection of visit type data at the time of the clinical encounter. Implementing this change would not only provide clearer insights but also potentially eliminate the necessity for using a predictive model, as accurate and structured data capture would be readily available. In addition, the model performance was dependent on the annotations provided by veterinary experts. Our interobserver agreement results, with a Fleiss Kappa of 0.91, underline the consistency among expert annotators, providing a reliable basis for the model’s training. The reliance of the model’s training on the agreement among veterinarians in classifying visits underscores the pivotal role of expert judgment in this process, especially as during the training process the annotating veterinarians could not identify the visit type for 926 visits. It also brings to light the limitations of the model in situations where there is a lack of consensus among experts. To ensure practical relevance, future efforts should aim to integrate this aspect into routine workflow and use more varied data for model training. Additionally, our sample was skewed towards dogs (78.2% of visits), which may have influenced the model’s performance across species. Future research should investigate further refining the model’s ability to differentiate between wellness and non-wellness visits, especially in cases where expert consensus might be challenging. In particular, the discrepancy in wellness visit classification between cats and dogs calls for a more nuanced approach to species-specific care patterns in future model development.
Wellness and preventive care visits are crucial in the veterinary-client-patient relationship, offering a chance to inform clients and detect underlying diseases early. The classification of wellness visits in large-scale RWE data could expand research on the role preventive care plays in the identification of subclinical disease. While the utilization of a visit classification model on retrospective RWE data may expedite the assessment of the clinical value of preventive care visits, further prospective studies are essential to validate these findings.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: the datasets presented in this article are not readily available because the data used in this study contains identifiable transaction and medical history level information. Requests to access these datasets should be directed to [email protected] .
Author contributions
DS: Writing – review & editing, Writing – original draft, Visualization, Validation, Project administration, Methodology, Investigation, Formal analysis, Conceptualization. MC: Writing – review & editing, Supervision, Investigation, Data curation, Conceptualization. JR: Writing – review & editing, Project administration, Data curation. KK: Writing – review & editing, Software, Data curation, Conceptualization. DM: Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization. DK: Writing – review & editing, Supervision, Software, Investigation, Data curation, Conceptualization.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
DS, MC, JR, KK, DM, and DK involved in this study were employees of IDEXX Laboratories.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1348162/full#supplementary-material
SUPPLEMENTAL APPENDIX 1 | Initial Annotator Training Performance
SUPPLEMENTARY APPENDIX 2 | Model Calibration and additional performance metrics.
1. Min, K-D. Scoping review of machine learning and deep learning algorithm applications in veterinary clinics: situation analysis and suggestions for further studies. J Vet Clin . (2023) 40:243–59. doi: 10.17555/jvc.2023.40.4.243
Crossref Full Text | Google Scholar
2. Estrada, AH, Spake, A, Kleman, ME, Leeder, D, Blischok-Lapekas, D, Margiocco, M, et al. Diagnostic accuracy of computer aided electrocardiogram analysis in dogs. J Small Anim Pract . (2021) 62:145–9. doi: 10.1111/jsap.13267
3. Solomon, J, Bender, S, Durgempudi, P, Robar, C, Cocchiaro, M, Turner, S, et al. Diagnostic validation of vertebral heart score machine learning algorithm for canine lateral chest radiographs. J Small Anim Pract . (2023). 64:769–775. doi: 10.1111/jsap.13666
4. ICD-11. Available at: https://icd.who.int/en
Google Scholar
5. Lustgarten, JL, Zehnder, A, Shipman, W, Gancher, E, and Webb, TL. Veterinary informatics: forging the future between veterinary medicine, human medicine, and one health initiatives—a joint paper by the Association for Veterinary Informatics (AVI) and the CTSA one health Alliance (COHA). JAMIA Open . (2020) 3:306–17. doi: 10.1093/jamiaopen/ooaa005
6. Pineda, AL, Bear IV, GR, Venkataraman, AM, Sandeep Ayyar, Z, Page, RL, Bustamante, CD, et al. Deep learning facilitates rapid cohort identification using human and veterinary clinical narratives. bioRxiv . (2018) bioRxiv:429720. doi: 10.1101/429720
7. Davies, H, Nenadic, G, Alfattni, G, Arguello Casteleiro, M, al Moubayed, N, Farrell, SO, et al. Text mining for disease surveillance in veterinary clinical data: part one, the language of veterinary clinical records and searching for words. Front Vet Sci . (2024) 11:1352239. doi: 10.3389/fvets.2024.1352239
8. DeepTag: inferring diagnoses from veterinary clinical notes. NPJ Digital Medicine. Available at: https://www.nature.com/articles/s41746-018-0067-8 .
9. American Animal Hospital Association. Preventive care pet health resources. AAHA. Available at: https://www.aaha.org/practice-resources/pet-health-resources/preventive-care/
10. Janke, N, Coe, JB, Bernardo, TM, Dewey, CE, and Stone, EA. Use of health parameter trends to communicate pet health information in companion animal practice: a mixed methods analysis. Vet Rec . (2022) 190:e1378. doi: 10.1002/vetr.1378
11. AAHA-AVMA canine preventive healthcare guidelines. American Veterinary Medical Association. Available at: https://www.avma.org/resources-tools/avma-policies/aaha-avma-canine-preventive-healthcare-guidelines .
12. Preventive Health Care Guidelines for Cats. VCA Animal Hospitals. VCA. Available at: https://vcahospitals.com/know-your-pet/preventive-health-care-guidelines-for-cats .
13. Rodan, I, and Sparkes, AH. Preventive health Care for Cats. The Cat . (2012):151–80. doi: 10.1016/B978-1-4377-0660-4.00008-9
14. Willems, A, Paepe, D, Marynissen, S, Smets, P, van de Maele, I, Picavet, P, et al. Results of screening of apparently healthy senior and geriatric dogs. J Vet Intern Med . (2017) 31:81–92. doi: 10.1111/jvim.14587
15. Dell’Osa, D, and Jaensch, S. Prevalence of clinicopathological changes in healthy middle-aged dogs and cats presenting to veterinary practices for routine procedures. Aust Vet J . (2016) 94:317–23. doi: 10.1111/avj.12481
16. Paepe, D, Verjans, G, Duchateau, L, Piron, K, Ghys, L, and Daminet, S. Routine health screening: findings in apparently healthy middle-aged and old cats. J Feline Med Surg . (2013) 15:8–19. doi: 10.1177/1098612X12464628
17. Jeffery, U, Jeffery, ND, Creevy, KE, Page, R, and Simpson, MJ. Variation in biochemistry test results between annual wellness visits in apparently healthy Golden retrievers. J Vet Intern Med . (2021) 35:912–24. doi: 10.1111/jvim.16021
18. Matsumoto, M, and Nishimura, T. Mersenne twister: a 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans Model Comput Simul . (1998) 8:3–30. doi: 10.1145/272991.272995
19. R Development Core Team. R: A language and environment for statistical computing . Vienna, Austria: R Foundation for Statistical Computing (2009).
20. Fryda, T., LeDell, E., Gill, N., Aiello, S., Fu, A., Candel, A., et al. h2o: R Interface for the ‘H2O’ scalable machine learning platform (Version 3.44.0.3). CRAN . (2023). Available at: https://cran.r-project.org/package=h2o
21. Friedman, JH. Greedy function approximation: a gradient boosting machine. Ann Stat . (2001) 29:1189–232. doi: 10.1214/aos/1013203451
22. Walter, SD, and Irwig, LM. Estimation of test error rates, disease prevalence and relative risk from misclassified data: a review. J Clin Epidemiol . (1988) 41:923–37. doi: 10.1016/0895-4356(88)90110-2
23. Dawid, AP, and Skene, AM. Maximum likelihood estimation of observer error-rates using the EM algorithm. J R Stat Soc . (1979) 28:20–8.
24. Harrell, F. HMISC: Harrell Miscellaneous . (2021).
25. Harrell, F. rms: Regression Modeling Strategies (Version 6.8-1). CRAN . Available at: https://cran.r-project.org/package=rms (2021).
26. Wickham, H.RStudio. Tidyverse: Easily install and load the ‘Tidyverse’ (Version 2.0.0). CRAN . Available at: https://cran.r-project.org/package=tidyverse (2017).
27. Wickham, H., Chang, W., Henry, L., Takahashi, K., Wilke, C., Woo, K., et al. ggplot2: Create elegant data Visualisations using the grammar of graphics . Springer (2018).
Keywords: wellness visit, machine learning, preventative care, clinical visit, wellness
Citation: Szlosek D, Coyne M, Riggott J, Knight K, McCrann DJ and Kincaid D (2024) Development and validation of a machine learning model for clinical wellness visit classification in cats and dogs. Front. Vet. Sci . 11:1348162. doi: 10.3389/fvets.2024.1348162
Received: 01 December 2023; Accepted: 05 August 2024; Published: 30 August 2024.
Reviewed by:
Copyright © 2024 Szlosek, Coyne, Riggott, Knight, McCrann and Kincaid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald Szlosek, [email protected]
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
| Property | Value |
|---|---|
| Status | |
| Version | |
| Ad File | |
| Disable Ads Flag | |
| Environment | |
| Moat Init | |
| Moat Ready | |
| Contextual Ready | |
| Contextual URL | |
| Contextual Initial Segments | |
| Contextual Used Segments | |
| AdUnit | |
| SubAdUnit | |
| Custom Targeting | |
| Ad Events | |
| Invalid Ad Sizes |
Login to your account
If you don't remember your password, you can reset it by entering your email address and clicking the Reset Password button. You will then receive an email that contains a secure link for resetting your password
If the address matches a valid account an email will be sent to __email__ with instructions for resetting your password
Current Issue Links
Articles in press.
- Past Issues
Submit Mobile
- Submit article
- Call For Papers
- Press Releases Opens in new window
- Submit Article Opens in new window
- Aims and Scope
- For Authors
- Supports Open Access Opens in new window
- Alerts Opens in new window
- ARPAS Opens in new window
Applied Animal Science Is Now a Gold Open Access Journal
As of january 2024, applied animal science will be fully transitioning to a gold open access journal, offering authors wider visibility and readers unrestricted access to critical advancements in animal science research., this month's editor's choice, invited review: role for isoacids in dairy nutrition*.
- Firkins et al.
Perspective and Commentary: Use of soy-based feedstuffs in low-alfalfa, high–corn silage diets for dairy cows
Producer perceptions of us livestock indemnity policy.
- Campbell et al.
Identifying and quantifying key sustainability indicators for pastoral dairy-beef production systems
- Kearney et al.
More from Applied Animal Science
Mapping the Key Sustainability and Profitability Metrics in Ireland’s Dairy-Beef Production Systems

Livestock Producers Prioritize Fair Market Value in New Study of Indemnity Policy Preferences

Understanding the Role of Branched-Chain Volatile Fatty Acids in Enhancing Dairy Cow Nutrition and Performance

Soy-Based Feedstuffs for Low-Alfalfa, High-Corn Silage Dairy Cow Diets
Applied Animal Science
Applied Animal Science (AAS) is a peer-reviewed, Gold open access scientific journal and the official publication of the American Registry of Professional Animal Scientists (ARPAS). This journal publishes scientific discoveries and applications for the animal sciences and animal production systems. The primary focus is on innovations and technologies that have immediate (or emerging) application in domestic animal agriculture. More fundamental (basic) articles are also welcome if the application to animal science is readily obvious and can be communicated to readers. Types of articles published include reviews, invited reviews, articles from symposia and conferences, original research articles, short communications, and technical notes.
About ARPAS
The purpose of the American Registry of Professional Animal Scientists (ARPAS) is to provide certification of professional status for qualified members of the society, to strengthen animal sciences among the professions, and to promote animal sciences and the profession of animal scientists. Continual education is required of all certified professionals to keep abreast of rapidly changing technology in their fields.

ARPAS members and nonmembers are invited and encouraged to submit manuscripts for review and possible publication in Applied Animal Science (AAS) . The main purpose of AAS is to publish high quality, peer-reviewed, science-based information for animal industry professionals on a wide range topics and species. More information [PDF]
- Submit a Manuscript Opens in new window
Editorial Team

David K. Beede Editor-in-Chief Read Dr. Beede's biography

Eric van Heugten Associate Editor Read Dr. van Heugten's biography

Daniel Rivera Associate Editor Read Dr. Rivera’s biography

Kristin Hales Associate Editor Read Dr. Hales' biography
Most Read (Last 30 Days)
Effects of backgrounding-phase rate of gain on performance and carcass characteristics of feedlot steers.
- Blom et al.
Effects of feeding lubabegron fumarate or ractopamine hydrochloride to beef × dairy crossbred steers raised under small-pen commercial feedlot conditions in western Canada
- Shreck et al.
REVIEW: Updated scientific evidence on the welfare of gestating sows kept in different housing systems
- John J. McGlone
Invited Review: Examples and opportunities for artificial intelligence (AI) in dairy farms*
- De Vries et al.
R eview : Milk production from dairy cows in Argentina: Current state and perspectives for the future
- Lazzarini et al.
Most Cited (Previous 3 Years)
- Articles & Issues
- Current Issue
- List of Issues
- Collections
- Editor's Choice
- In the News
- Press Releases
- Call for Submissions
- Instructions for Authors
- Tips for Creating Tables in Microsoft Word
- Figure Preparation Guidelines
- Submit a Manuscript
- Open Access License
- Permissions
- Submission/Revision Checklist
- Researcher Academy
- Journal Info
- About the Journal
- About Open Access
- ARPAS Membership
- Contact Information
- Editorial Board
- New Content Alerts
- Reviewer Appreciation
- Activate Online Access
- Subscriptions and Pricing
The content on this site is intended for healthcare professionals.
- Privacy Policy
- Terms and Conditions
- Accessibility
- Help & Contact


Virtual Tour
Experience University of Idaho with a virtual tour. Explore now
- Discover a Career
- Find a Major
- Experience U of I Life
More Resources
- Admitted Students
- International Students
Take Action
- Find Financial Aid
- View Deadlines
- Find Your Rep

Helping to ensure U of I is a safe and engaging place for students to learn and be successful. Read about Title IX.
Get Involved
- Clubs & Volunteer Opportunities
- Recreation and Wellbeing
- Student Government
- Student Sustainability Cooperative
- Academic Assistance
- Safety & Security
- Career Services
- Health & Wellness Services
- Register for Classes
- Dates & Deadlines
- Financial Aid
- Sustainable Solutions
- U of I Library

- Upcoming Events
Review the events calendar.
Stay Connected
- Vandal Family Newsletter
- Here We Have Idaho Magazine
- Living on Campus
- Campus Safety
- About Moscow

The largest Vandal Family reunion of the year. Check dates.
Benefits and Services
- Vandal Voyagers Program
- Vandal License Plate
- Submit Class Notes
- Make a Gift
- View Events
- Alumni Chapters
- University Magazine
- Alumni Newsletter

SlateConnect
U of I's web-based retention and advising tool provides an efficient way to guide and support students on their road to graduation. Login to SlateConnect.
Common Tools
- Administrative Procedures Manual (APM)
- Class Schedule
- OIT Tech Support
- Academic Dates & Deadlines
- U of I Retirees Association
- Faculty Senate
- Staff Council
Department of Animal, Veterinary and Food Sciences
Department of Animal, Veterinary & Food Sciences
University of Idaho
Physical Address: E. J. Iddings Agricultural Science Laboratory, Rm 213 606 S Rayburn St
Mailing Address: 875 Perimeter Drive MS 2330 Moscow, ID 83844-2330
Phone: 208-885-6345
Fax: 208-885-6420
Email: [email protected]
Web: uidaho.edu/cals/avfs
Robert J. Collier
Department head, amin ahmadzadeh, associate professor — meat science, jacob w. bledsoe, assistant professor & extension specialist — aquaculture research, carolyn hovde bohach, distinguished professor, mireille chahine, professor & extension specialist — dairy, gwinyai chibisa, associate professor — ruminant nutrition & metabolism, lauren christensen, assistant professor — mixed practice production medicine, michael colle, joseph dalton, stacey doumit, senior instructor, melinda ellison, associate professor & extension specialist — range livestock & sheep, daniel fitzsimons, assistant professor, benton glaze, professor & extension specialist — beef, professor & extension specialist — beef; cattle management lead, rinker rock creek ranch, janna hamlett, clinical faculty & extension specialist — food processing, denise konetchy, associate professor — small ruminant medicine, vikas kumar, assistant professor — fish nutrition & nutrigenomics, mark mcguire, university distinguished professor, scott minnich, brenda murdoch, associate professor, amy skibiel, associate professor — lactation physiology, jim sprinkle, izabelle teixeira, assistant professor & extension specialist — dairy, james vanleuven, matt doumit, senior associate dean & director, academic programs.
Ag Science, Room 213
208-885-9489
Animal, Veterinary and Food Sciences University of Idaho 875 Perimeter Drive MS 2330 Moscow, ID 83844-2330

View Full Profile
College of Agricultural and Life Sciences
Ph.D., University of Illinois, 1976 M.S., Eastern Illinois University, 1973 B.S., Eastern Illinois University, 1969
Ag Science, Room 219
208-885-7409

Ph.D., Virginia Tech M.S., Virginia Tech B.S., Delaware Valley
- AVS 172: Principles and Practices of Dairy Science
- AVS 222: Animal Reproduction and Breeding
- AVS 472: Dairy Cattle Management
- AVS 475: Advanced Dairy Management
Ag Science, Room 221
208-885-0990

Phil applies industry experience to teach, advise students and others involved in the Pacific Northwest meat production and processing community; conducts applied research in meat quality with emphasis in beef.
Ph.D., Colorado State University, 2009 M.S., California Polytechnic State University — San Luis Obispo, 2006 B.S., California Polytechnic State University — San Luis Obispo, 2004
- AVS 109: The Science of Animals that Serve Humanity
- AVS 363: Animal Products for Human Consumption
- AVS 499: Advanced Butchery
Hagerman Fish Culture Experiment Station
208-837-9096 x1105
Hagerman Fish Culture Experiment Station 3059F National Fish Hatchery Rd. Hagerman, ID 83332

Jake works to improve economic and environmental sustainability of aquaculture production through applied research (fish microbiome, nutritional physiology, disease, genetics and water quality). Extension priorities included workforce development and marketing.
Ph.D., University of Idaho, 2019 M.S., Southern Illinois University, 2015 B.S., Purdue University, 2012 A.S., Purdue University, 2012
Mines Building, Room 317
208-596-1747

Carolyn's focus is infectious disease with emphasis on the foodborne pathogen E. coli O157:H7. Primary interests are in the relationship of this human pathogen with healthy cattle.
Ph.D., University of Minnesota, 1985 MT (ASCP), Swedish Hospital Medical Center, 1976 B.S., University of Illinois, 1975
- FS 517: Scientific Writing
- FS 518: Oral Seminar
208-736-3609
uidaho.edu/cals/twin-falls
Twin Falls Research and Extension Center University of Idaho 315 Falls Avenue, Evergreen Bldg. Twin Falls, Idaho 83301

Ag Science, Room 212
208-885-7074

Ph.D., University of Saskatchewan, 2013 M.Sc., University of Saskatchewan, 2008 B.Sc., University of Zimbabwe, 2005
- AVS 305: Animal Nutrition
- AVS 306: Feeds and Ration Formulation
- AVS 474: Beef Cattle Science
- AVS 517: Macronutrient Metabolism
Ag Biotech, Room 119
208-885-9703

Lauren teaches undergraduate courses at U of I and WSU vet student rotations, giving students hands-on learning experiences to develop, integrate, and apply critical thinking and technical skills.
M.S., University of Idaho, 2022 DVM, Kansas State University, 2017 B.S., California Polytechnic State University-San Luis Obispo, 2013
- AVS 267: Anatomy and Physiology of Domestic Animals
- AVS 268: Companion Animal Diseases
- AVS 317: Artificial Insemination and Pregnancy Detection
- Mixed large animal and production medicine clinical instructor, WSU senior veterinary students
Food Research Center, Suite 103
208-885-6007

Ph.D., University of Idaho, 2017 M.S., University of Idaho, 2014 B.S., University of Wisconsin-River Falls, 2011
- AVS 110: Science of Animal Husbandry
- AVS 110 Laboratory: Science of Animal Husbandry
- CORS 232: Science on Your Plate
- AVS 263: Live Animal and Carcass Evaluation
- AVS 274: Beef Feedlot Systems
- FS 489: Food Product Development
- AVS 463/563: Growth and Lactation
Caldwell Complex
208-454-7633
uidaho.edu/cals/caldwell
Caldwell Research and Extension Center University of Idaho 1904 E Chicago St, Suite A, B Caldwell, ID 83605

Dalton’s research is focused on increasing the efficiency of artificial insemination in dairy cattle, including sperm dosage, heat detection accuracy, synchronization programs, AI technician proficiency and identification of fertility markers.
Ph.D., Virginia Tech M.S., Utah State B.S., Cal Poly
Ag Science, Room 209
208-885-7143

M.Sc., Michigan State University, 1994 B.Sc., Michigan State University, 1989
- AVFS 101: Intro to Animal, Vet and Food Sciences
- AVS 466: Equine Science and Management
- AVS 450: Issues in Animal Agriculture
- AGLS 494: CALS Peer Leaders
208-756-2749
uidaho.edu/cals/nmcreec
Nancy M. Cummings Research, Extension & Education Center University of Idaho 16 Hot Springs Ranch Road Carmen, ID 83462

Ellison's research focuses primarily on the effects of grazing livestock on wildlife and range.
Ph.D., University of Wyoming, 2016 M.S., University of Wyoming, 2013 B.S., University of Wyoming, 2011
Ag Biotech, Room 307
208-885-5054

My research focuses on identifying the molecular processes that underlie the regulation of cardiac contraction and relaxation in the context of normal and diseased states, such as hypertrophic cardiomyopathy and heart failure.
Ph.D., University of California, Irvine, 1989 B.S., University of California, Irvine, 1983
- AVFS 371/373: Anatomy and Physiology
208-736-3638
Twin Falls Research and Extension Center University of Idaho 315 Falls Avenue, Evergreen Bldg. Twin Falls ID 83301

Benton focuses on breeding and genetics, beef quality assurance, general management and reproduction. He serves as a resource for beef producers, allied industries, veterinarians, Extension educators and the general public.
Ph.D., Kansas State University M.S., University of Missouri-Columbia B.S., Tarleton State University
- AVS 330: Genetics of Livestock Improvement
- AVS 474: Beef Cattle Science
Nancy M. Cummings Research Extension & Education Center University of Idaho 16 Hot Springs Road Carmen, ID 83462

The center is the University of Idaho’s primary cow/calf and forage research center. John’s program conducts applied research to enhance food production and sustainability of beef production operations.
208-731-9363

Janna works and assists the local food processing industry through TechHelp, UI Extension and the Animal, Veterinary and Food Sciences Department.
Ed.D., Idaho State University, 2021 M.S., Washington State University, 2016 B.S., University of Idaho, 2003
Ag Science, Room 216
208-885-7390
konetchy@uidaho

Owner and clinical practitioner of a rural mixed animal veterinary practice. The practice emphasized strategies, of preventative medicine, herd health and client education. In addition I was a temporary instructor at the University of Idaho.
MPH, University of Iowa, 2017 DVM, Washington State University, 1986 B.S., Washington State University, 1984 B.S., Washington State University, 1983
- AVS 452: Physiology of Reproduction
- AVS 471: Animal Disease Management
- AVS 476: Sheep Science
- Small ruminant medicine clinical instructor — WSU senior veterinary students
Ag Science, Room 214 or Aberdeen center
208-885-1088
uidaho.edu/cals/aberdeen

Kumar’s research interest revolves around aquaculture (marine and freshwater); nutrition, physiology and genomics of aquatic animals. His primary research is to develop nutritionally balanced, environmentally sound and cost-effective seafood production.
Ph.D., University of Hohenheim, Germany, 2011 M.F.Sc., Central Institute of Fisheries Education, Mumbai, India, 2006 B.F.Sc., Central Agricultural University, India 2004
- AVS 504: Fish Nutrition
- AVS 404: Fish Nutrition
- AVFS 204: Anatomy & Physiology of Domesticated Animals
- AVS 501: Graduate Seminar
- FISH 600: Research and Dissertation
Languages other than English spoken
Ag Science, Room

Ag Biotech, Room 209
208-310-2477

Scott's focus is on bacterial pathogenesis of Yersinia enterocolitica and Y. pestis , etiologic agents of yersiniosis and bubonic plague respectively; and ability to suppress the innate immune response of mammalian hosts.
Ag Biotech, Room 311
208-885-2088

Murdoch’s research focuses on characterizing the relationship between genetic variation in mammals and traits which are valued and important to society.
Ph.D., University of Alberta B.Sc., University of Alberta
Ag Biotech, Room 303
208-885-1161

Skibiel’s research interests lie at the intersection of lactation biology, maternal effects and performance. These topics are studied using observational and experimental approaches and from both evolutionary and dairy production perspectives.
Ph.D., Auburn University, 2012 M.S., Auburn University, 2007 B.S., Juniata College, 2003
- AVS 463: Growth and Lactation (fall 2019)

Ph.D., Texas A & M University M.S., Montana State University B.S., Brigham Young University A.S., Ricks College
208-736-3604

Teixeira’s research focuses on improving our understanding of nutrient utilization and animal efficiency to reduce the impact of ruminant production on the environment and enhance sustainability of ruminant production systems.
Ph.D., Sao Paulo State University, Brazil, 2004 M.S., Sao Paulo State University, Brazil, 2000 B.S., Federal University of Lavras, Brazil, 1997
IRIC, Room 333
Animal, Veterinary and Food Sciences University of Idaho 875 Perimeter Drive MS 1122 Moscow, ID 83844-1122

Ag Science, Room 65
208-885-7984
Academic Programs College of Agricultural and Life Sciences 875 Perimeter Drive MS 2336 Moscow, ID 83844-2336

Academic Programs
Ph.D., Michigan State University, East Lansing, 1994 M.S., South Dakota State University, 1989 B.S., Washington State University, 1987
Staff support
Robin inman, administrative coordinator, jessica christensen, student support specialist, kevin carnahan, senior researcher, lucelia de moura pereira, research specialist, ryan foster, laboratory services manager, logan harper, dairy center manager, organic group leader, talia holtmeyer, meat lab assistant, mina mahdavi-yekta, james nasados, meat science lab manager, barbara nielsen, research support scientist, makayla proett, beef feedlot manager, anup tuladhar, laboratory coordinator/manager, janet williams, senior research scientist, kelci scharff, dairy center assistant manager.
Ag Biotech, Room 111
208-885-6345

Ag Science, Room 123
208-885-2030

B.S., Utah State University, 2012
Ag Science, Room 123E
208-885-6831

Ph.D., Washington State University M.S., Brigham Young University B.S., University of Central Missouri
Ag Biotech, Rooms 213, 316, 322
208-310-5793

Provide training for lab personnel and support departmental and laboratory activities; oversight for proper maintenance and operation of instruments; and generate data for abstracts and manuscripts for publication.
Ph.D., Federal University of Parana, 2022 M.S., Federal University of Parana, 2016 B.S., Maringa State University, 2014
Holm Veterinary Science Center
208-885-7466
Analytical Sciences Laboratory University of Idaho 875 Perimeter Drive MS 2203 Moscow, ID 83844-2203
Analytical Sciences Laboratory
208-885-1152
Palouse Research, Extension and Extension Center University of Idaho 875 Perimeter Dr. MS 2090 Moscow, ID 83844-2090

Palouse Research, Extension and Education Center
B.S., University of Idaho, 2018

Ph.D., University of Idaho B.S., University of Wyoming
Vandal Brand Meats
208-885-6727
Animal, Veterinary and Food Sciences University of Idaho 875 Perimeter Drive MS 2330 Moscow, Idaho 83844-2330
Assistant to the Meat Lab manager in processing, slaughter of beef, pork and lamb.
B.S., Washington State University, 2021
Ag Science, Room 129B

M.S., Korea University, 2017 B.S., Dongduk Women’s University, 2015
Ag Science, Room 217C, lab 214
323-806-2085

Mina’s focus is on relationships between nutrient metabolism and various measures of animal productivity, animal product quality and safety. The interactions between lipid-soluble vitamins, nutritional characteristics, safety aspects of animal-based products.
Ph.D., Tehran, Iran, 2019 M.S., Tehran, Iran, 2014 B.S., Tehran, Iran, 2010


541-290-8237

B.S., University of Idaho, 2021
208-885-6275

Ph.D., University of Idaho, 2021 M.S., University of Idaho, 2019
Ag Biotech, Room 305
208-885-6351; 208-874-3213 (c)

Ph.D., University of Idaho, 2016 M.S., Oklahoma State University, 1987 B.S., Oklahoma State University, 1985
Dairy Center Office
208-885-3526
Palouse Research, Extension and Education Center University of Idaho 875 Perimeter Drive MS 2090 Moscow, ID 83844-2090

Palouse Research Extension and Education Center
B.S., University of Idaho, 2022
Faculty emeritus
- Gulhan Unlu , professor, food microbiology
- Pedram Rezamand , professor, dairy nutrition
- James England, professor, former veterinarian for Nancy M. Cummings Research, Extension and Education Center
- Rick Norell, professor, Extension dairy specialist
- Gordon Murdoch, professor
- Ron Hardy , professor and former director of the Aquaculture Research Institute
- Andrzej J. Paszcynski , professor, former UI-WSU School of Food Science
- Context Statement pdf
- Promotion and Tenure Guide pdf
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Springer Nature - PMC COVID-19 Collection

CRISPR–Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing
Irina gostimskaya.
The University of Manchester, M1 7DN Manchester, UK
The development of a method for genome editing based on CRISPR–Cas9 technology was awarded The Nobel Prize in Chemistry in 2020, less than a decade after the discovery of all principal molecular components of the system. For the first time in history a Nobel prize was awarded to two women, Emmanuelle Charpentier and Jennifer Doudna, who made key discoveries in the field of DNA manipulation with the CRISPR–Cas9 system, so-called “genetic scissors”. It is difficult to overestimate the importance of the technique as it enables one not only to manipulate genomes of model organisms in scientific experiments, and modify characteristics of important crops and animals, but also has the potential of introducing revolutionary changes in medicine, especially in treatment of genetic diseases. The original biological function of CRISPR–Cas9 system is the protection of prokaryotes from mobile genetic elements, in particular viruses. Currently, CRISPR–Cas9 and related technologies have been successfully used to cure life-threatening diseases, make coronavirus detection tests, and even to modify human embryo cells with the consequent birth of babies carrying the introduced modifications. This intervention with human germplasm cells resulted in wide disapproval in the scientific community due to ethical concerns, and calls for a moratorium on inheritable genomic manipulations. This review focuses on the history of the discovery of the CRISPR–Cas9 system with some aspects of its current applications, including ethical concerns about its use in humans.
A HISTORY OF THE DISCOVERY OF THE MAIN COMPONENTS OF THE CRISPR–Cas9 SYSTEM
CRISPR – clustered regularly interspaced short palindromic repeats – were first discovered in the sequences of DNA from Escherichia coli bacteria and described in 1987 by Ishino et al. [ 1 ] from Osaka University (Japan). At that time sequencing of these difficult-to-study DNA fragments took several months, but neither their origin nor their significance in the bacterial cell were understood by their discoverers. Although in the early work in this field, the biological function of the CRISPR system had not yet been elucidated, scientists had already proposed a way to use the information encoded in CRISPR loci in medical research, namely, for genotyping various strains of bacteria: initially on Mycobacterium tuberculosis [ 2 ], and later on Streptococcus pyogenes [ 3 ]. As it turned out, CRISPR loci had a high degree of polymorphism in different strains of the same species of pathogenic bacteria, which enabled the identification of bacterial strains in clinical conditions.
A significant breakthrough in understanding the biological function of CRISPR loci occurred with the discovery of Francisco Mojica of the University of Alicante (Spain), who came across similar structures in the archaeal genome of Haloferax mediterranei in 1995 [ 4 ]. Their presence in two evolutionarily remote domains of life suggested these elements’ great functional significance, and served as an impetus for further research. Mojica noticed the similarity of the elements he described in archaea with previously found DNA repeats in bacterial genomes, and was one of the first scientists to hypothesize that these unusual loci include fragments of foreign DNA and are, in fact, a part of the immune system of bacteria and archaea [ 5 ]. In the same year as Mojica, two other laboratories independently reached similar conclusions [ 6 , 7 ], announcing the beginning of an era of active research into this extraordinary natural phenomenon. In line with the theory of the prokaryotic immune system, viral DNA fragments (“spacers” 17-84 bases long), separated by short palindromic repeats (23-50 bases [ 8 ]) and grouped into clusters in intergenic regions, represent a library of potentially dangerous genetic information (for an overview of the microbial antiviral arsenal, see reviews by Isaev et al. [ 9 , 10 ]). Initially, it was assumed that such a system would work by the mechanism of RNA interference. However, in the publication of Marraffini and Sontheimer, it was experimentally demonstrated for the first time that the actual target of the immune system of prokaryotes was foreign DNA [ 11 ], and not mRNA, and, therefore, the use of such a system in the laboratory could represent a potential tool for genomic editing. Interestingly, later studies demonstrated that some of the described CRISPR systems do work with RNA molecules directly [ 12 , 13 ] and, therefore, can be used to deactivate specific transcripts inside the cell in a selective way [ 14 , 15 ].
The first experimental information about the mechanism of action of the CRISPR system was obtained in 2007 in the studies of two French food scientists, Rodolphe Barrangou and Philippe Horvath, who worked with yoghurt cultures of bacteria Streptococcus thermophilus for the Danish company Danisco [ 16 ]. Due to the company’s rich collection of bacterial strains collected since the 1980s, scientists have been able to trace the historical course of the bacterial acquisition of spacers at the CRISPR locus in response to viral attacks by bacteriophages. The addition of new spacers in this work caused acquired immunity to the corresponding new types of bacteriophages in S. thermophilus : observation which subsequently led to the authors obtaining one of the first patents in this area [ 17 ] and the start of bacterial cultures’ “vaccination” with the use of CRISPR-based technology by Danisco in 2005 [ 18 ].
Currently, CRISPR repeats have been found in most archaeal genomes and nearly half of the studied bacterial ones, but they have not been found in eukaryotic or viral DNA sequences. The existence of CRISPR repeats in mitochondria was suggested in one of the earliest publications on the subject (the same article described CRISPR in cyanobacteria for the first time) [ 19 ]. The authors used a set of previously published data on the sequencing of mitochondrial plasmids from Vicia faba L. beans [ 20 ], and their conclusions were further cited by Mojica et al. [ 21 ], but these observations were not confirmed in later studies [ 8 ].
At the time of initial discoveries, a variety of different acronyms was used for CRISPR by individual scientific groups, which presently complicates the search for early articles on the topic. The current name for CRISPR first appeared in Jansen et al. [ 22 ] in 2002 and was suggested by Mojica in correspondence between the two collaborating scientific groups. The same publication was the first one to describe the presence of genes associated with CRISPR repeats (named by the authors cas1-4 , CRISPR-associated genes). These genes were found in close proximity to the CRISPR loci of various prokaryotes, and two of them contained motifs characteristic of helicase and nuclease, which supported the authors’ hypothesis about the non-random association of the cas genes with the CRISPR locus, and their involvement in DNA metabolism. Also in 2002, the same neighborhood of genes was described by a team of scientists led by Eugene Koonin from the NCBI Institute (Bethesda, USA), but the association of these genes with CRISPR arrays was not discerned by them at the time [ 23 ]. From the moment of the first discovery of genes associated with the CRISPR system, to the present day, their truly extraordinary abundance and diversity have been found in prokaryotic cells, including representatives of the families of helicases, nucleases, polymerases, and others. Proteins associated with this system can be assigned to either the adaptive module (participating in the acquisition of immunity, main representatives – Cas1 and Cas2), or the effector module (directly involved in the destruction of mobile genetic elements through their recognition and cleavage), with some additional and regulatory proteins also found to be associated with the system [ 24 ]. At present, a way of classification is recognized in which all currently known CRISPR–Cas systems are divided into 2 classes and 6 types, which, in turn, are also divided into numerous subtypes: at the time of writing the review, Makarova et al. [ 25 ] described >30 subtypes ( Fig. 1 ). The main difference between the classes is that the effector module of Class 1 systems is represented by a complex of several proteins, while in Class 2 it is a single multidomain protein (Cas9, Cas12, or Cas13) [ 26 - 28 ].

Conventional classification of known CRISPR–Cas systems.
Of all the known Cas proteins, the most studied ones are the proteins belonging to the system of directional cutting of foreign DNA (and, as it was found out later, in some cases, RNA), the so-called “genetic scissors”, among which is the nuclease Cas9. This protein was first described in connection with its association with CRISPR repeats in an article by Bolotin et al. [ 6 ], where it was originally named Cas5 (other alternative names are Csn1 and Csx12). In addition, the authors identified the presence of the HNH motif (His-Asn-His), which is also found in other nucleases. Another important observation made by Bolotin et al. was the discovery of a specific pattern in the nucleotide sequences on one side of the described spacers of the CRISPR arrays, but the understanding of the role for this phenomenon was only revealed in later studies. Currently, short motifs adjacent to protospacers but absent in the original spacers of the CRISPR locus are called PAMs (protospacer adjacent motifs) [ 29 ]. Protospacers are DNA fragments that are attacked by the immune system of prokaryotes, and are identical to the corresponding spacers at the CRISPR locus, except for the PAM motif. These motifs are important at the stage of recognition of potentially dangerous genetic information; their presence at the end of the sequence signals that the DNA fragment is foreign and needs to be destroyed, while the DNA sequences stored in the CRISPR locus as spacers and not containing PAM motifs are not attacked by the prokaryotic immune system.
A crucial player in the CRISPR–Cas9 system turned out to be a short RNA molecule, a processed product of transcription from the CRISPR locus that directs proteins of the prokaryotic immune system to foreign molecules with genetic information. A group of researchers led by John van der Oost (Wageningen University, the Netherlands), who described the existence of such RNA molecules, gave them the name crRNA (CRISPR-associated RNA). It was also noted that the initial result of transcription from the CRISPR locus is a pre-crRNA precursor molecule consisting of several spacers and repeats, which is later cleaved into individual fragments [ 30 ]. In the work of the group led by Virginijus Siksnys (Vilnius University, Lithuania), it was demonstrated that the length of the actual “guide” crRNA sequence of 20 base pairs, complementary to the target DNA, is necessary and sufficient for the nuclease activity of the CRISPR–Cas complex, even if the spacer in CRISPR locus is represented by a longer sequence of nucleotides [ 31 ]. This publication was one of two in vitro studies, carried out in parallel and independently in competing laboratories, that described, for the first time, how the Cas9 enzyme uses crRNA to attack foreign DNA.
The final missing piece in the puzzle, without which it is impossible to assemble a working CRISPR–Cas9 system in vitro , turned out to be another short RNA molecule, discovered in connection with its participation in crRNA processing by Emmanuelle Charpentier’s group in 2011 [ 32 ]. This molecule, essential for nuclease activity, was named tracrRNA (trans-activating CRISPR RNA). In subsequent work, ultimately acknowledged by the Nobel Prize, the role of tracrRNA in the mechanism of target DNA cutting was shown. It was also proposed at the time that two RNA molecules, crRNA and tracrRNA, could be combined into one chimeric molecule (sgRNA – single guide RNA), which greatly facilitated the practical use of the CRISPR–Cas9 system in subsequent applications [ 33 ]. Figure 2 shows the timeline of the historical events in the discovery of the CRISPR–Cas9 system’s components: initially the CRISPR locus itself, then the proteins associated with it, including Cas9, and later, two RNA molecules necessary for the formation of the ribonucleoprotein complex and recognition of substrate DNA.

Historical timeline of discoveries of the components of the CRISPR–Cas9 system. 1987 – Short DNA repeats, later called CRISPR, were first noticed in bacterial genomes, and, in 1995, also found in archaea. 2005 – The role of CRISPR loci in the protection of prokaryotes from foreign genetic information was proposed, and the Cas9 protein was described for the first time (initial information on proteins associated with the CRISPR locus appeared in 2002). Two RNA molecules, crRNA and tracrRNA, were discovered as part of the complex in 2007 and 2011, respectively. The Nobel Prize-winning work, where all of the components were assembled in vitro and two RNA molecules combined into one strand for the ease of use of the system, was published in 2012.
USE OF THE CRISPR–Cas9 SYSTEM IN EUKARYOTIC CELLS
The discovery of the necessary and sufficient components of the CRISPR–Cas9 system started a race to be the first to apply the system to the genetic editing of human and animal cells. In January 2013, almost simultaneously, five research articles authored by different research teams appeared, all reporting that they had achieved the goal. Two publications from the same issue of the journal Science , offering probably the best approach to the problem had been produced by the laboratories of George Church (Harvard University, USA) and Feng Zhang (Broad Institute, USA). In these publications, it was shown that for successful DNA editing in human cells, it was necessary to carry out several steps: these include codon optimization and the addition of a nuclear localization signal to the cas9 gene, lengthening of the sgRNA molecule (to improve the efficiency of the system), as well as the possible addition of a DNA template for homologous recombination with which the cells can repair the DNA double break (the last step was described only by the group of G. Church) [ 34 , 35 ]. Also in January 2013, similar publications came out from the laboratories of Jennifer Doudna (Berkeley College, USA) [ 36 ], Jin-Soo Kim (Seoul University, South Korea) [ 37 ] and J. Keith Joung (Harvard School of Medicine, USA) [ 38 ]. In the last article [ 38 ], the described work was carried out on zebrafish rather than human cells but, importantly, the use of the CRISPR–Cas9 system on germline cells was demonstrated for the first time.
FIRST CRYSTALLOGRAPHIC STUDIES
The most studied protein from the Cas group is the Cas9 nuclease; in the ~20 years since the discovery of the cas genes more than 20,000 articles in the PubMed system mention the name Cas9 in one context or another. Attempts to obtain detailed information about the structure of this protein resulted in the first two crystallographic studies being published almost simultaneously: in February 2014 two crystal structures of Cas9 appeared in the database PDBe (“Protein Data Bank in Europe”), and the accompanying articles were published in the journals Nature and Cell [ 39 , 40 ]. The structure that came out of the laboratory of Jennifer Doudna was of an apo-protein (PDBe ID 4cmp, PDBe DOI: 10.2210/pdb4cmp/pdb), while the research group of Osamu Nureki (University of Tokyo, Japan) succeeded in crystallising the protein in a complex with a “guide”-RNA and “target”-DNA (PDBe ID 4oo8, PDBe DOI: 10.2210/pdb4oo8/pdb).
These, as well as many subsequent studies, used the Cas9 protein from S. pyogenes , SpCas9, which consists of 1368 amino acids and is a multidomain and multifunctional endonuclease. Crystal structures revealed that the Cas9 protein is spatially divided into 2 lobes: a target recognition lobe and a nuclease lobe, with the guide RNA and target DNA occupying the positively charged groove at their interface. The key structures of the nuclease lobe of SpCas9 are 2 domains: HNH and RuvC, each of them cleaves one of the target DNA strands. Figure 3 shows the general architecture of the SpCas9–sgRNA–DNA complex, where the complex secondary structure of the bound RNA molecule, and the unwound state of the double-stranded DNA molecule with the formation of a DNA–RNA heteroduplex can be seen (PDB ID 5F9R, PDB DOI: 10.2210/pdb5F9R/pdb, [ 41 ]). At the time of writing, hundreds of crystal structures of the Cas9 family proteins are available from the PDB, PDBe, and PDBj databases.

Three-dimensional organization of the Cas9 protein in the complex with “guide” RNA (sgRNA) and substrate (Target DNA), crystallographic data (PDB ID 5F9R, PDB DOI: 10.2210/pdb5F9R/pdb).
PATENT DISPUTE
The understandable motive of individual scientists, as well as organizations involved in the study of the CRISPR–Cas9 system, was the possible financial gain potentially obtainable from the use of this promising technology. One of the first patent applications was filed jointly by the University of California at Berkeley, representing Doudna, the University of Vienna (where one of the two lead authors from the key publication on CRISPR–Cas9 worked [ 33 ]), and Charpentier as an individual inventor in accordance with the rules of the University of Umeå (Sweden), where Charpentier worked at the time of publication of the article [ 18 ]. This patent application was filed in May 2012 [ 42 ], while in December 2012 Zhang and the Broad Institute also submitted a patent application [ 43 ] simultaneously with the acceptance of Zhang’s paper on human cells’ editing for publication in Science [ 35 ]. Initially, it was Zhang’s application that turned out to be successful and resulted in a patent in April 2014, while Doudna’s application was still pending at that time. Doudna’s team disagreed with the decision, after which a long dispute between the two parties followed, including appeals and court hearings which ultimately led to an ambiguous situation in CRISPR–Cas9 licensing. Due to the fact that by 2019 both competing parties had patents in this area, some of the biotech companies that used the CRISPR–Cas9 system on human cells received a license from the team of Doudna, while others – from Zhang. However, the U. S. Patent and Trademark Office Appeal Board in February 2022 again confirmed the priority of Zhang and the Broad Institute in the position of the patent holder for the use of CRISPR–Cas9 in human cells, which caused disappointment and frustration from the opposing side, and financial complications for companies licensed by the team of Doudna [ 44 ]. Doudna and Charpentier, however, won a similar dispute in Europe, and also hold major patents on the use of technology in the U.K., China, Japan, Australia, New Zealand, and Mexico [ 18 ].
GENE THERAPY AND ETHICAL ISSUES ASSOCIATED WITH IT
The haste with which competing laboratories sought to bring their research to the public’s attention, as well as the race to patent this technology, were indicators of the significance of this scientific breakthrough. Undoubtedly, one of the main driving forces that motivated many scientists to take part in research using this particular technology was the potential of modifying human cells, both somatic and germline. However, despite the apparent advantages of the CRISPR–Cas9 system, numerous ethical and technical difficulties stand in the way of researchers who dream of curing life-threatening diseases, especially if the genetic changes resulting from such manipulations can be inherited.
Gene therapy was administered for the first time in September 1990: a four-year-old girl suffering from adenosine deaminase (ADA) deficiency received an infusion of genetically engineered T-lymphocytes. Cells taken from the girl’s blood were modified using a viral vector – a deactivated virus that carries a healthy copy of the gene. As journalists who covered the story noted “rarely in modern medicine has an experiment been filled with so much hope”, and the doctor who performed this procedure, W. French Anderson, became known as the “father of gene therapy”. As time went on, however, the disturbing evidence of the adverse side effects of some attempts at gene therapy in both animals and humans began to accumulate. The tragic story of Jesse Gelsinger, an American teenager from Philadelphia who died from the effects of gene therapy in 1999, shocked the world and caused widespread skepticism and a significant delay in the development of the technology. In the case of Gelsinger, a large-scale autoimmune response of the body to a viral vector carrying the ornithine transcarbamylase gene led to a sharp increase in body temperature, renal and pulmonary failure, jaundice, impaired blood clotting, and subsequent death within only four days from the moment of gene therapy administration [ 45 ].
Extensive discussions of the safety and, importantly, the ethical issues arising from the possibility of potential gene therapy with CRISPR–Cas9 began soon after the first publications showing this system’s use in human cells. One of the first steps in initiating formal discussions was taken by Doudna, who organized a conference on scientific, medical, legal, and ethical issues related to the genomic modification, held in the Napa Valley in California in January 2015. A subsequent report of the results of the conference was published in March 2015 in the journal Science [ 46 ], which essentially carried recommendations to strongly discourage work on introducing heritable changes in human embryonic cells, at least for the duration of active discussions of the social, environmental and ethical consequences of such manipulations. Almost simultaneously with this report, a comment was also published in the journal Nature about the serious risks linked to creating heritable changes in human embryos [ 47 ]. The authors expressed concerns that premature work on embryonic cells could have a negative impact on the field of gene therapy in general, and could set back the work of researchers attempting to treat genetic and infectious diseases in somatic cells for years. The March 2015 report from the Napa conference and the commentary in Nature urging not to edit the human embryonic genome were released amidst growing agitation in the scientific community over leaked news that such experiments had actually already been carried out. A group of scientists from Sun Yat-sen University (Guangzhou, China), after unsuccessful attempts to get their manuscript accepted by the journals Nature and Science , in April 2015 finally published their article on the use of the CRISPR–Cas9 system on human embryonic cells [ 48 ]. The researchers emphasized that they used non-viable embryos obtained by the fusion of two sperm cells with one egg and, therefore, discarded by in vitro fertilization (IVF) laboratories. The main conclusion of the article was that the CRISPR–Cas9 technology at the time of the study was not yet ready for use on human embryonic cells due to the identified shortcomings in the system’s efficiency and specificity. A comment of the journal Protein & Cell (Beijing, China), that published this work, stated that the article (in addition to its scientific value) would promote an open exchange of information about current research in the area; and despite the ambiguity of the issue and conflicting opinions on the topic, the publication would stimulate the necessary discussions about genomic editing of germline cells. Interestingly, the manuscript had been sent to Protein & Cell together with the references obtained during previous attempts to publish the work, and was accepted by the editors for publication within two days from the date of submission. The subsequent debate in the scientific community was described as “epic” [ 49 ] and provoked interest in this complex issue from the wider public, as well as in governmental and regulatory organizations in various countries.
The notorious scandals caused by the conduct of medical experiments on humans in the past have led to the creation of general international guidelines on bioethics. The best-known documents in this area are the Nuremberg Code, developed after the trial of Nazi doctors in 1947, and the subsequent Declaration of Helsinki from 1964, which expanded the principles of the code and detailed the application of these principles to clinical research. Another important document, the Belmont Report, was issued by the National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research in the United States in 1978. This commission was created in the wake of shocking revelations of an inhumane syphilis study from 1932 to 1972 in Tuskegee. For decades, hundreds of impoverished African-American men infected with syphilis have been studied for the progression of their disease. Although penicillin had become the standard treatment for syphilis by 1947, it was not offered to study participants, despite the obvious physical suffering of the patients and the continued spread of the infection in their families.
The Nuremberg Code, the Declaration of Helsinki and the Belmont Report are based on the basic ethical principles of biomedical research, such as respect for the individual, informed consent of the patient, understanding of the risks and benefits, voluntary participation, fairness in the conduct of experiments, maximum professionalism of the researchers, etc. These principles, and their application in medical practice, are relevant to the events of November 2018, when the Chinese scientist Jiankui He announced the birth of babies who, for the first time, had undergone gene modification using the CRISPR–Cas9 system. The injection of this system into the mother’s egg was made at the stage of the IVF procedure immediately after the fusion of the sperm, and therefore all the changes potentially introduced into the genome during this procedure would be heritable. The world scientific community was shocked at how premature such medical experiments were, and the high degree of risk taken by the researchers conducting the experiment. In particular, scientists were worried about the possibility of creating unplanned (“off-target”) mutations in the genome of future babies. At the time of the experiment He (also known under the shortened name JK – from Jiankui) was not a well-known figure in the CRISPR–Cas9 community, however, after the announcement of his experiments, he attracted world-wide attention. He studied physics at the University of Science and Technology (Hefei, China) and then moved to the United States, where he received his PhD under the supervision of Michael Deem, Professor of Physics, Astronomy and Bioengineering at Rice University (Houston, Texas), and later worked as a post-doc at Stanford University (California) in the laboratory of Professor Stephen Quake. In the group of Deem He used the methods of theoretical biophysics, mathematical modelling and computer simulations, publishing papers on, among other things, influenza virus strains and spacer sequences in CRISPR loci [ 50 , 51 ], while in the laboratory of Quake, he learned the methods of molecular biology and became interested in the innovative technologies of Silicon Valley. Returning to China, He continued his collaboration with Deem, and also successfully implemented the innovative ideas in the field of DNA sequencing of his second supervisor, Quake, creating a successful company Direct Genomics based on the technology [ 18 , 52 ]. In China, he became quite famous as a young scientist and successful entrepreneur who had returned from abroad under the Thousand Talents program. He received a position and a laboratory at the Southern University of Science and Technology (SUStech, Shenzhen), and participated in the creation of several start-up companies [ 53 ]. The next step in his career resulted in the biggest medical scandal of the last decade. In 2017 on WeChat social media platform, He announced that he was recruiting volunteers from among married couples who wanted to produce children genetically modified to be resistant to the human immunodeficiency virus (HIV). Among the conditions of recruitment was that in the couple who wished to participate in the experiment both people had a university degree, so that they had enough educational background to understand the basics of science and medicine. A second condition was for the man to be HIV-positive and for the woman – HIV-negative: a situation in which the risk of transmitting the virus to the baby would be minimal (provided that the sperm was “washed” during the IVF procedure), but made it likely that the couple’s motivation to participate in the experiment would be high [ 53 ]. He planned to modify the CCR5 gene, a known receptor on the cell surface, through binding to which the human immunodeficiency virus enters the cell. About 300 people responded to the advertisement, of these, 20 couples were selected for the next round of consultations, during which the participants learned about the procedure and the possible risks. From these consultations 11 couples agreed to participate in the studies, of which seven were ultimately selected by the researchers for the next stage – the IVF procedure with an additional step of genome editing. The motivation of individual participants was, apparently, not only the possibility of having children (the IVF procedure in China is prohibited if one of the parents has HIV infection), but also the desire to take part in an “historic” experiment designed to benefit future generations [ 53 ]. Ultimately, after several unsuccessful attempts, from a selected group of participants 2 pregnancies led to the birth of babies who had undergone a genomic modification procedure using the CRISPR–Cas9 system. Quite a lot is known about the first pregnancy, which resulted in the birth of two twin girls, Lulu and Nana (pseudonyms used in the press and scientific literature in order to protect their identity). Very little information is available on the second pregnancy, which resulted in the birth of another child. Since this event occured after the scandal caused by the birth of the first twins, many details of the second pregnancy remained a secret. A manuscript written by He, based on the results of the first pregnancy and named “Birth of twins after genome editing for HIV resistance” remains unpublished, but has been leaked to the scientific community [ 54 , 55 ]. It has become known, for example, that in one of the embryos both copies of CCR5 were inactivated (Nana), while in the second, only one was modified (Lulu) [ 56 ]. Therefore, only Nana has a chance to be protected from HIV infection in the future, at least from the main variants of the virus that enter the cell through binding to the CCR5 receptor. In the case of Lulu, unfortunately, the treatment will provide no protection, since one copy of the CCR5 gene is enough to produce the corresponding receptor on the membrane. It is believed that two embryos were implanted in the uterus of a future mother in the hope that at least one of them will lead to the birth of a genetically modification baby. The twins were born premature (at 31 weeks) and spent the first weeks of their lives in neonatal incubators but were otherwise described as “healthy” [ 53 ]. Scientists who had gained access to the unpublished manuscript of He, also noted that several cells selected for sequencing early in embryonic development were in fact mosaics, an observation that led to increased criticism of He’s work. In the case of mosaicism, any information obtained during the sequencing of selected cells cannot be extrapolated to the entire embryo as a whole. Therefore, at the time of the key decision of whether to transfer the embryos into the womb, the researchers could not be sure that the CRISPR–Cas9 system did not produce any dramatic off-target mutations in the remaining cells of the embryos, even if the sequencing results showed the absence of such modifications in the selected cells. Many other aspects of the conduct of the study also received harsh criticism from the scientific and medical community [ 54 ], including the questionable circumstances of obtaining permission from the ethics committee of a hospital in Shenzhen, the level of qualification of He for clinical research (lack of medical education and adequate experience in the field), the choice of the gene that has undergone editing (social rather than medical reasons for patients seeking help), possible side effects from the lack of a valid copy of CCR5 , etc. According to an American cardiologist and Professor of Medicine at the University of Pennsylvania Kiran Musunuru, the first babies of “the CRISPR generation”, unfortunately, were born not as a result “of a historic scientific achievement, but rather a historic ethical fiasco” [ 56 ]. A preceding PR-campaign conducted by He and his team resulted in fairly flattering initial news coverage of his work in the People’s Daily (the largest newspaper group in China). However, the following international scandal led to the placement of He under house arrest, and then to a 3-year prison sentence. He has already been released from prison, but little is known about his whereabouts and future plans [ 57 ].
A few months after the described scandal the Russian scientist Denis Rebrikov stirred up the international scientific community with a statement about his intention to become the second scientist in the world to create genetically modified babies. Rebrikov, a Professor at the Pirogov Russian National Research Medical University and Head of the Laboratory of Genomic Editing at the Center for Obstetrics, Gynecology and Perinatology, announced that his research facility was potentially ready to transfer modified embryos into the mother’s womb in June 2019 [ 58 ]. As in the experiments of He, he was planning to edit the CCR5 gene, and the preliminary work from his laboratory on non-viable embryos was published in the Bulletin of the Russian National Research Medical University [ 59 ]. The reaction of the scientific community to the statement was heated and primarily negative. In October 2019 the journals Nature and Science published news feeds reporting that at that time, Rebrikov had already switched to editing the GJB2 gene associated with inherited deafness, and was in the process of selecting couples who would agree to take part in the experiment [ 60 , 61 ]. However, in numerous interviews with journalists Rebrikov emphasized that he would only conduct such experiments after obtaining all necessary permits from both regulatory and ethical authorities. This significantly distinguished his approach from He’s, who informed the scientific community about the birth of babies with a modified genome post factum . The Ministry of Health of the Russian Federation (following the recommendation of the World Health Organisation) later made a statement that the decision to grant permission for such a study would be premature and irresponsible, which prevented the further development of the situation at least until the situation in the regulatory sphere changes [ 62 ].
At the time of writing this review, the state of the legal framework that regulates the issue of genomic editing of human embryonic cells varies greatly in different countries. Thus, genomic modification of embryos for purposes other than reproductive is allowed in at least 11 countries, including China, the U.S., and the U.K. Nineteen countries, including Belarus, Canada, Sweden, and Switzerland, prohibit such experiments. Many other countries (Russia among them) take an intermediate or indeterminate position. The situation with the introduction of inherited genomic changes into embryos subsequently used for reproductive purposes is even more complicated [ 63 ].
MEDICAL APPLICATIONS WITH HUMAN SOMATIC CELLS
Despite increased attention to the introduction of heritable changes in germline cells, the less controversial and currently more common use of CRISPR–Cas9 for medical purposes is the modification of human somatic cells. As described above, in the first attempts at gene therapy (1990) an adeno-associated viral vector was used that delivered a healthy copy of the gene into cells (in the U.S. this technology was finally approved for clinical use only in 2017 [ 64 ]). The next step in the development of gene therapy was the introduction of genomic editing with the use of Homing Endonucleases (HEs), Zinc Fingers Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and later also CRISPR–Cas9 [ 65 ]. The first human clinical studies using CRISPR–Cas9 commenced in October 2016 in China [ 66 ]. The PD-1 gene was inactivated ex vivo in blood cells in the hope that such modified cells, would attack the non-small-cell lung cancer that the patient suffered from when returned to circulation. In the U.S., ex vivo therapy using CRISPR–Cas9 was first performed in July 2019 on a patient with sickle cell anemia (CRISPR Therapeutics, founded by Charpentier). The therapy significantly improved the patient’s condition for at least a few months after the procedure, however the cost of such treatment at the time of its implementation in the United States was estimated to be in the region of 0.5-1.5 million U.S. dollars. The high current cost of CRISPR–Cas9 therapy will probably act as an obstacle to its widescale use, even if clinical trials confirm the efficacy and safety of such treatment [ 18 ]. Currently, the most expensive drug on the market is Zolgensma, another gene therapy treatment used for spinal muscular atrophy ($2.125 million per dose). Zolgensma directly delivers a working copy of the defective gene into cells with the use of adeno-associated virus, a method different from genomic editing using nucleases [ 67 ].
The first example of an in vivo clinical study in which cells undergo in situ genomic editing with nucleases was performed using the ZFNs technology. Sangamo Therapeutics first performed this procedure in July 2017 on a patient suffering from Hunter syndrome (a rare genetic disease, form of mucopolysaccharidosis). The pioneers in using CRISPR–Cas9 for in vivo genomic editing were Editas Medicine (March 2020) [ 68 ]. A drug called EDIT-101 was injected locally into the retina of a patient suffering from a form of inherited blindness caused by a mutation in the CEP290 gene. Currently, various clinical studies are underway on the use of CRISPR–Cas9 for the treatment of diseases such as Alzheimer’s disease, various types of cancers, high cholesterol, angioedema, acute myeloid leukemia, and even androgenetic alopecia (baldness). Another promising application for CRISPR–Cas9 in the future could be the treatment of infectious diseases caused by such pathogens as, for example, HIV and human papillomavirus [ 65 ].
CONCLUSIONS
The discovery of CRISPR–Cas9 as an immune system in prokaryotes at the turn of the 20th-21st centuries – a finding at first glance only relevant to microbiology – has led to a revolution in the field of genomic manipulations. New opportunities have opened up in multiple areas of biomedicine, such as molecular diagnostics of infectious and non-infectious diseases (e.g., genotyping of bacterial strains, detection of viruses, and identification of genetic mutations in circulating extracellular DNA in patients with lung cancer [ 69 ]), as well as in the development of a potentially new method of immunization, DNA vaccines [ 18 ]. One of the more unusual examples of the application of the CRISPR–Cas9 system was the cultivation of brain-like organelles carrying different variants of the important NOVA1 gene characteristic of modern humans, Neanderthals, and Denisovans [ 70 ]. The development of CRISPR–Cas9 technology is a good example of how discoveries made in the course of basic research can change entire fields of science and technology, expanding the horizons of the possible. This ground-breaking technique is a worthy continuation of such exciting scientific events as the publication of the double-stranded structure of DNA by Watson and Crick in 1953, the birth of the first child by in vitro fertilization in 1978, and the cloning of Dolly the sheep in 1996. In the coming years the scientific community will watch with interest the development of legislation and ethical principles in the application of the CRISPR–Cas9 system in genome editing, as well as in what other areas of science this promising technology will find its application.
Acknowledgments
The author recalls with warmth and gratitude the years spent in the laboratory of Andrei Dmitrievich Vinogradov at the Department of Biochemistry of Moscow State University. The experiments conceived by Andrei Dmitrievich invariably brought interesting results, while his vast knowledge in various fields of science enabled staff and students to feel confident that any questions would be answered, and the time spent in the laboratory would bring well-deserved results. The publication of the results of the work carried out under the supervision of Andrei Dmitrievich gave the author the necessary start in scientific life and the opportunity to continue research in other laboratories and other fields of knowledge. A unique team of scientists, selected by Andrei Dmitrievich: Vera Georgievna Grivennikova, Tatiana Vadimovna Zharova, and Eleonora Vladimirovna Gavrikova, provided a family atmosphere of trust and support in the laboratory, for which the author is very grateful.
Abbreviations
| CRISPR-associated genes | |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| crRNA | CRISPR-associated RNA |
| PAM | protospacer adjacent motif |
| sgRNA | single guide RNA |
| Cas9 | Cas9 protein from |
| tracrRNA | trans-activating CRISPR RNA |
Ethics declarations
The author declares no conflicts of interest. This article does not contain a description of the studies performed by the author with the participation of people or animals as objects.
INVITRO is a provider of full Central laboratory services and Clinical trial management services for Phase I-IV.
INVITRO Laboratory also performs Local laboratory services for International clinical trials. Because of our broad experience and specialized expertise, we’re in unique position to support safety laboratory tests as an alternative to the different site bases.

Local laboratory services are offered for Russia, Belarus and Kazakhstan
INVITRO Clinical Research can use its own Courier logistics service for pick-up and delivery of samples to the laboratory.
Due to the absence of custom between these countries, we are one of the small groups of organization with the capability and expertise to conduct Laboratory services and development projects for either local or global clinical trials.
All the tests for Clinical trials are performed in the Moscow laboratory.
It is stocked with high-tech equipment and located in a space of 3 500 sqm including:
- 365 work days per year, including monitoring and registration of biological samples around the clock
- Quality assurance of laboratory services in accordance with ISO 9001, ISO 15189 certificates
- Validated methodologies, logistical and data management support, reliable and flexible reporting systems and cohesive quality control, which are critical to meet the requirements of your organization and regulatory agencies.
Our Project teams perform ongoing monitoring and proactive communications to provide high-quality outcomes. Protocol specifications are built in during database setup to provide customized reports and sophisticated result flagging for monitoring. Our secure Internet-based, remote data access system allows members of the project team to review lab results in real time. INVITRO scientists, technologists and project managers can assist with protocol development to provide the most appropriate tests for each study, and all INVITRO staff is trained and certified accordingly. INVITRO Laboratory offers the flexibility required for quick and customized responses to unique client needs for any size project.
We strongly believe that a collaborative relationship between INVITRO Laboratory and Clinical Investigators is fundamental study success.
We provide:
- Design of study-specific manual
- Lab Kit manufacturing in accordance with GLP requirements
- Study-specific requisition forms and courier documents
- Biomaterial transportation in compliance with IATA requirements
- Telephone assistance
- Specimen handling
- Long-Term Storage and frozen specimen tracking
- Personal Account of clinical trials – an opportunity to use a tool that allows you to monitor the results of laboratory research, query, order a courier and lab kits online and have access to laboratory documents and project -specific documents
INVITRO clinical research
- Patient service
- Project Management
- Request for proposal
Invitro Clinical Research
INVITRO Clinical Research has supported clinical study teams at pharma companies and CROs facing the challenges of recruiting study subjects
www.invitro-cr.com
- General Information
- Quality Assurance
- Request for Proposal
Legal Disclaimer
- Privacy Policy
- Cookie Policy

COMMENTS
About Journal Clinical Research in Animal Science is a peer-reviewed scholarly journal dedicated to the advancement in all fields of animal sciences. CRAS is a platform to advance the knowledge and to improve Animals health and are undergone with clinical trials to find new drug to alleviate pain. CRAS mainly focuses on providing an international forum to professionals, Researchers and other ...
For all potential issues concerning the description of the publication identified by this bibliographic record (missing or wrong data etc.), please contact the ISSN National Centre mentioned above by clicking on the link. Display linked data. ISSN 2770-6729 (Online) | Clinical research in animal science.
Animal Science Journal. With over 90 years of long publishing history, Animal Science Journal provides a forum for presenting research in all fields of animal and poultry science: genetics and breeding, genetic engineering, reproduction, embryo manipulation, nutrition, feeds and feeding, physiology, anatomy, environment and behavior, animal ...
Novel veterinary approaches to herd health: From laboratory diagnostics to treatment This Special Issue welcomes full research articles, systematic reviews, case studies, and randomized controlled trials showcasing novel approaches to veterinary diagnostic services and treatment in diverse herd health management systems.
Section Information Animals is an international peer-reviewed scientific open access journal that publishes original research articles, reviews, communications, and short notes. Animals is characterized by numerous sections that offer substantial new insight into different subject areas of animal science. The section of Veterinary Clinical Studies includes a great variety of Veterinary ...
Read more about Crimson Publishers-Clinical Research in Animal Science. CTAS is a peer-reviewed scholarly journal dedicated to the advancement in all fields of animal sciences. CRAS is a platform ...
The animal model deals with the species other than the human, as it can imitate the disease progression, its' diagnosis as well as a treatment similar to human. Discovery of a drug and/or component, equipment, their toxicological studies, dose, side effects are in vivo studied for future use in humans considering its' ethical issues.
The journal encourages the submission of high-quality novel research that has clear implications for the prevention, treatment, or control of zoonotic and animal diseases, including improved understanding of disease pathogenesis and epidemiology, and that therefore contribute to a substantial improvement of animal and human health. Papers studying the origin, pathogenesis, and spread of ...
High-Impact Research from Journal of Animal Science Explore a collection of the most read and most cited articles making an impact in Journal of Animal Science published within the past two years. This collection will be continuously updated with the journal's leading articles so be sure to revisit periodically to see what is being read and cited.
Open science has become a buzzword in the scientific community that too often misses the practical application for individual researchers. This Essay, provides a guide to choosing the most appropriate tools to make animal research more transparent.
Clinical veterinary research that falls within the definition of 'non-experimental veterinary practice' and is conducted on client-owned animals is not subject to ethical review under either European or UK legislation. In other words, in Europe (including the UK), the laws regulate the use of laboratory animals for scientific procedures ...
Journal Rankings on Animal Science and Zoology. Agricultural and Biological Sciences (miscellaneous) Biochemistry, Genetics and Molecular Biology (miscellaneous) Business, Management and Accounting (miscellaneous) Economics, Econometrics and Finance (miscellaneous) Organizational Behavior and Human Resource Management.
Part of the third most-cited veterinary science journal, this section explores animal medicine, animal disease, and a comparative approach to structure, diagnosis, therapy and prevention.
The resulting evidence suggests that the collective harms and costs to humans from animal experimentation outweigh potential benefits and that resources would be better invested in developing human-based testing methods. Keywords: animal research, medical testing, human health, human ethics, drug development, animal ethics Go to:
This article discusses the ethical issues and principles involved in animal experimentation, and provides examples and recommendations for researchers.
Developments in the reporting of clinical signs will enhance the scientific value gained from animal experiments and further address the ethical cost. This paper discusses systematic approaches to the observation and reporting of clinical signs in animals (to be) used for research.
Medical procedures necessary for routine care can induce stress in both the veterinary and human clinical situations. In the research environment, nonhuman primates undergo procedures like physical examination, blood sampling, and intravenous drug or fluid administration either as a part of routine veterinary care or during the modeling of clinical disease and interventions under study ...
Nonhuman animals have long been and continue to be routinely used in biomedical and behavioral research to promote human health. When SARS-CoV2 infections triggered a race to develop and scale global access to vaccines in 2019, two key innovations happened to the supply chain of nonhuman animals created, raised, and used for science: (1) experiments and trials regarded as essential were ...
Journal Rankings on Animal Science and Zoology. Agricultural and Biological Sciences (miscellaneous) Biochemistry, Genetics and Molecular Biology (miscellaneous) Business, Management and Accounting (miscellaneous) Economics, Econometrics and Finance (miscellaneous) Organizational Behavior and Human Resource Management.
Materials and methods: The algorithm was initially trained using a Gradient Boosting Machine model with a dataset of 11,105 clinical visits from 2012 to 2017 involving 655 animals (85.3% dogs and 14.7% cats) across 544 U.S. veterinary practices. Three validators were tasked with classifying 400 visits, including both wellness and other types of ...
A clinical research requires a systematic approach with diligent planning, execution and sampling in order to obtain reliable and validated results, as well as an understanding of each research methodology is essential for researchers. Indeed, selecting ...
Applied Animal Science. Applied Animal Science (AAS) is a peer-reviewed, Gold open access scientific journal and the official publication of the American Registry of Professional Animal Scientists (ARPAS). This journal publishes scientific discoveries and applications for the animal sciences and animal production systems.
The Department of Animal, Veterinary and Food Sciences conducts focused, comprehensive and integrated research and teaching programs that directly support animal agriculture and food science. The animal and veterinary science bachelor's degree offers four options: business, dairy, pre-veterinary and production.
Browse the University of Idaho Department of Animal, Veterinary and Food Sciences faculty profiles for useful information on current research interests and projects, as well as course information.
The discovery of the necessary and sufficient components of the CRISPR-Cas9 system started a race to be the first to apply the system to the genetic editing of human and animal cells. In January 2013, almost simultaneously, five research articles authored by different research teams appeared, all reporting that they had achieved the goal.
Laboratory. INVITRO is a provider of full Central laboratory services and Clinical trial management services for Phase I-IV. INVITRO Laboratory also performs Local laboratory services for International clinical trials. Because of our broad experience and specialized expertise, we're in unique position to support safety laboratory tests as an ...